| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,620,993 – 14,621,105 |
| Length | 112 |
| Max. P | 0.973671 |

| Location | 14,620,993 – 14,621,105 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 75.42 |
| Mean single sequence MFE | -29.27 |
| Consensus MFE | -16.11 |
| Energy contribution | -15.83 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.55 |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.973671 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
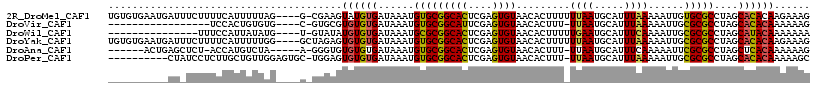
>2R_DroMel_CAF1 14620993 112 - 20766785 UGUGUGAAUGAUUUCUUUUCAUUUUUAG----G-CGAAGUAUGUGAUAAAUGUGCGGCACUCGAGUGUAACACUUUUUUAAUGCAUUUAAAAAUUGUGCGCCUAGCACACAAGAAAG (((((((((((.......))))).((((----(-((..((((((.....)))))).((((....))))..(((.(((((((.....)))))))..))))))))))))))))...... ( -30.70) >DroVir_CAF1 338685 94 - 1 -----------------UCCACUGUGUG----C-GUGCGUGUGUGAUAAAUGUGCGGCAUUCGAGUGUAACACUUU-UUAAUGCAUUUAAAAAUUGCGCGCCUAGCACACAAAAAAG -----------------.....((((((----(-..(((((((........(((((......(((((...))))).-....)))))........)))))))...)))))))...... ( -29.09) >DroWil_CAF1 403790 97 - 1 ---------------UUUCCAUUAUAUG----U-GUAUAUGUGUGAUAAAUGCGCGGCACUCGAGUGUAACACUUUUUGAAUGCAUUUCAAAAUUGCGCGCCUAGCAUACAAAAAAA ---------------.............----.-.....((((((......(((((((((....))))......(((((((.....)))))))...)))))....))))))...... ( -22.90) >DroYak_CAF1 269361 113 - 1 UGUGUGAAUGAUUUCUUUUCAUUUUUGG----GCUAGAGUGUGUGAUAAAUGUGCGGCACUCGAGUGUAACACUUUUUUAAUGCAUUUAAAAAUUGCGCGCCUAGCACACAAGAAAG (((((((((((.......))))))....----(((((.(((((..((....(((..((((....))))..)))..((((((.....))))))))..)))))))))))))))...... ( -34.90) >DroAna_CAF1 301976 103 - 1 ------ACUGAGCUCU-ACCAUGUCUA-----A-GGGUGUGUGUGAUAAAUGUGCGGCACUCGAGUGUAACACUUU-UUAAUGCAUUUCAAAAAUUCGCGCCUAGCUCACAAAAAAG ------..((((((..-..........-----.-(((((((.((.......(((((......(((((...))))).-....))))).......)).)))))))))))))........ ( -23.95) >DroPer_CAF1 251543 105 - 1 ----------CUAUCCUCUUGCUGUUGGAGUGC-UGGAGUGUGUGAUAAAUGUGCGGCACUCGAGUGUAACACUUU-UUAAUGCAUUUAAAAAUUGCGCGCCUAGCACACAAAAAGC ----------..........(((.(((..((((-(((.(((((..((....(((..((((....))))..))).((-((((.....))))))))..)))))))))))).)))..))) ( -34.10) >consensus __________AU_UCUUUCCAUUUUUAG____C_GGGAGUGUGUGAUAAAUGUGCGGCACUCGAGUGUAACACUUU_UUAAUGCAUUUAAAAAUUGCGCGCCUAGCACACAAAAAAG .......................................((((((......(((((((((....)))).........((((.....))))......)))))....))))))...... (-16.11 = -15.83 + -0.28)
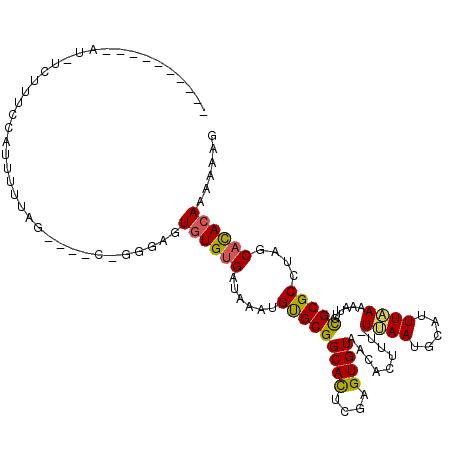

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:19:48 2006