| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,612,499 – 14,612,606 |
| Length | 107 |
| Max. P | 0.509909 |

| Location | 14,612,499 – 14,612,606 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 78.02 |
| Mean single sequence MFE | -30.41 |
| Consensus MFE | -19.29 |
| Energy contribution | -19.93 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.63 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.509909 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
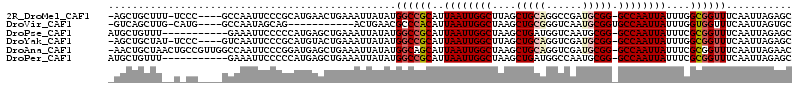
>2R_DroMel_CAF1 14612499 107 + 20766785 -AGCUGCUUU-UCCC----GCCAAUUCCCGCAUGAACUGAAAUUAUAUGGCCGCAUUAAUUGGCUUAGCUGCAGGCCGAUGCGG-GCCAAUUAUUUGGCGGUUUCAAUUAGAGC -....(((((-..((----(((((..((((((((..(((.........(((((.......)))))......)))..).))))))-)........)))))))........))))) ( -31.86) >DroVir_CAF1 328484 97 + 1 -GUCAGCUUG-CAUG----GCCAAUAGCAG-----------ACUGAACGCCCACAUUAAUUGGCUAAGCUGCGGGUCAAUGCGGUGCCAAUUAUUUGGUGGUUUCAAUUAGUGC -.((((..((-(((.----....)).))).-----------.))))....((((..((((((((...((((((......))))))))))))))....))))............. ( -29.60) >DroPse_CAF1 240229 102 + 1 AUGCUGUUU-----------GAAAUUCCCCCAUGAGCUGAAAUUAUAUGGCCGCAUUAAUUGGCUAAGCUGAUGGUCAAUGCGG-GCCAAUUAUUUCGCGGUUUCAAUUAGAGC ...(((.((-----------(((((((......))((.(((((....((((((((((.((..((...))..))....))))).)-))))...))))))).))))))).)))... ( -27.70) >DroYak_CAF1 260560 107 + 1 -AGCUGCUAU-UCCC----GUCAAUUCCCGCAUGUACUGAAAUUAUAUGGCCGCAUUAAUUGGCUUAGCUGCAGGUCGAUGCGG-GCCAAUUAUUUGGCGGUUUCAAUUAGAGC -.(((.(((.-..((----(((((.......(((((.......)))))((((((((..((..((......))..))..)))).)-)))......))))))).......)))))) ( -30.00) >DroAna_CAF1 288772 112 + 1 -AACUGCUAACUGCCGUUGGCCAAUUCCCGGAUGAGCUGAAAUUAUAUGGCAGCAUUAAUUGGCUAAGCUGCAGGUCGAUGCGG-GCCAAUUAUUUCGCGGUUUCAAUUAGAAC -((((((...(((((((..((..(((....)))..))..).......))))))...((((((((....(((((......)))))-))))))))....))))))........... ( -33.01) >DroPer_CAF1 242706 102 + 1 AUGCUGUUU-----------GAAAUUCCCCCAUGAGCUGAAAUUAUAUGGCCGCAUUAAUUGGCUAAGCUGAUGGCCAAUGCGG-GCCAAUUAUUUCGCGGUUUCAAUUAGAGC ...(((.((-----------(((((((......))((.(((((....(((((((((....((((((......)))))))))).)-))))...))))))).))))))).)))... ( -30.30) >consensus _AGCUGCUU__U_CC____GCCAAUUCCCGCAUGAGCUGAAAUUAUAUGGCCGCAUUAAUUGGCUAAGCUGCAGGUCAAUGCGG_GCCAAUUAUUUCGCGGUUUCAAUUAGAGC ................................................((((((..((((((((....(((((......))))).))))))))....))))))........... (-19.29 = -19.93 + 0.64)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:19:47 2006