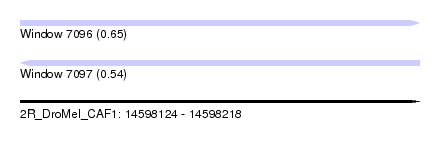
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,598,124 – 14,598,218 |
| Length | 94 |
| Max. P | 0.652838 |

| Location | 14,598,124 – 14,598,218 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 80.49 |
| Mean single sequence MFE | -18.68 |
| Consensus MFE | -12.93 |
| Energy contribution | -14.17 |
| Covariance contribution | 1.24 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.652838 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
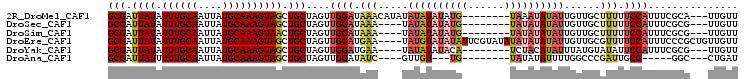
>2R_DroMel_CAF1 14598124 94 + 20766785 GCGAUUAUAUUUGCAAUUAUGCAAAGUAGCUGCUAGUUGGAUAAACAUAUAUAUAUAUG--------UAAAUAUAUUGUUGCUUUUUCCAUUUCGCA---UUGUU ((((.....((((((....))))))............((((.(((((...((((((...--------...))))))...)).))).))))..)))).---..... ( -18.20) >DroSec_CAF1 230206 90 + 1 GCGAUUAUAUUUGCAAUUAUGCAAAGUAGCUGCUAGUUGGAUAAA----UAUAUAUAUG--------UAUAUAUAUUGUUGCUUUUUCCAUUUCGCG---UUGUU ((((.....((((((....))))))(((((..((....))...((----(((((((...--------.))))))))))))))..........)))).---..... ( -17.80) >DroSim_CAF1 233540 90 + 1 GCGAUUAUAUUUGCAAUUAUGCAAAGUAACUGCUAGUUGGAUAAA----UAUAUAUAUG--------UAUAUAUAUUGUUGCUUUUUCCAUUUCGCG---UUGUU ((((.....((((((....))))))(((((..((....))...((----(((((((...--------.))))))))))))))..........)))).---..... ( -17.80) >DroEre_CAF1 237671 101 + 1 GCGAUUAUAUUUGCAAUUAUGCAAAGUAGCUGCUAGUUGGAUGAA----UAUGUAUAUAUUCGUAUAUAUAUAUAUUGUUGCGUUUUCCAUUUCCCGCUGUUGUU (((.((((.((((((....)))))))))).)))((((.(((..((----(((((((((((....)))))))))))))..((.(....)))..))).))))..... ( -22.20) >DroYak_CAF1 245793 90 + 1 GCGAUUAUAUUUGCAAUUAUGCAAAGUAGCUGCUAGUUGGAUGAA----UAUAUAUACA--------UCUACAUAUUUAUGUAUAUUCCAUUUCGCG---UUGUU (((.((((.((((((....)))))))))).)))..((..((((((----((((((((.(--------(......)).)))))))))).))))..)).---..... ( -19.50) >DroAna_CAF1 271101 82 + 1 GCGAUUAUUUUUGCAAUUAUGCAAAGUAGCUGCUAGUUGGAUAUC----GUUGU---UG--------UAUAUAUUUUGGCCCGAUUGCC-----GGC---CUGAU (((.(((((((.(((....)))))))))).)))..(((((..(((----(..((---((--------.........)))).))))..))-----)))---..... ( -16.60) >consensus GCGAUUAUAUUUGCAAUUAUGCAAAGUAGCUGCUAGUUGGAUAAA____UAUAUAUAUG________UAUAUAUAUUGUUGCUUUUUCCAUUUCGCG___UUGUU (((.((((.((((((....)))))))))).)))....((((.(((.....((((((((((......))))))))))......))).))))............... (-12.93 = -14.17 + 1.24)

| Location | 14,598,124 – 14,598,218 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 80.49 |
| Mean single sequence MFE | -15.80 |
| Consensus MFE | -7.04 |
| Energy contribution | -9.65 |
| Covariance contribution | 2.61 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.45 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.540750 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
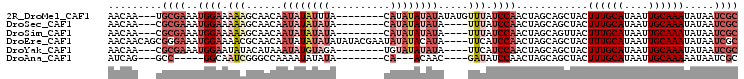
>2R_DroMel_CAF1 14598124 94 - 20766785 AACAA---UGCGAAAUGGAAAAAGCAACAAUAUAUUUA--------CAUAUAUAUAUAUGUUUAUCCAACUAGCAGCUACUUUGCAUAAUUGCAAAUAUAAUCGC .....---.((((..((((..(((((...((((((...--------...))))))...))))).))))............((((((....)))))).....)))) ( -17.80) >DroSec_CAF1 230206 90 - 1 AACAA---CGCGAAAUGGAAAAAGCAACAAUAUAUAUA--------CAUAUAUAUA----UUUAUCCAACUAGCAGCUACUUUGCAUAAUUGCAAAUAUAAUCGC .....---.((((..((((.........(((((((((.--------...)))))))----))..))))............((((((....)))))).....)))) ( -15.10) >DroSim_CAF1 233540 90 - 1 AACAA---CGCGAAAUGGAAAAAGCAACAAUAUAUAUA--------CAUAUAUAUA----UUUAUCCAACUAGCAGUUACUUUGCAUAAUUGCAAAUAUAAUCGC .....---.((((..((((.........(((((((((.--------...)))))))----))..))))....(((((((.......)))))))........)))) ( -15.80) >DroEre_CAF1 237671 101 - 1 AACAACAGCGGGAAAUGGAAAACGCAACAAUAUAUAUAUAUACGAAUAUAUACAUA----UUCAUCCAACUAGCAGCUACUUUGCAUAAUUGCAAAUAUAAUCGC .......((((....((((..........((((....))))..((((((....)))----))).))))............((((((....)))))).....)))) ( -15.40) >DroYak_CAF1 245793 90 - 1 AACAA---CGCGAAAUGGAAUAUACAUAAAUAUGUAGA--------UGUAUAUAUA----UUCAUCCAACUAGCAGCUACUUUGCAUAAUUGCAAAUAUAAUCGC .....---.((((..((((((((((((..........)--------)))))))...----....))))............((((((....)))))).....)))) ( -16.71) >DroAna_CAF1 271101 82 - 1 AUCAG---GCC-----GGCAAUCGGGCCAAAAUAUAUA--------CA---ACAAC----GAUAUCCAACUAGCAGCUACUUUGCAUAAUUGCAAAAAUAAUCGC ....(---(((-----.(....).))))..........--------..---....(----(((.......(((...))).((((((....))))))....)))). ( -14.00) >consensus AACAA___CGCGAAAUGGAAAAAGCAACAAUAUAUAUA________CAUAUAUAUA____UUUAUCCAACUAGCAGCUACUUUGCAUAAUUGCAAAUAUAAUCGC .........((((..((((.(((......((((((((..........)))))))).....))).))))............((((((....)))))).....)))) ( -7.04 = -9.65 + 2.61)
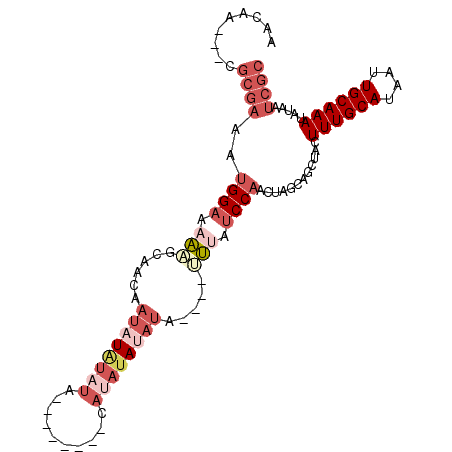
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:19:41 2006