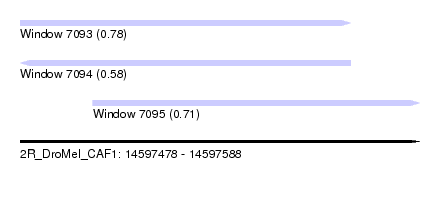
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,597,478 – 14,597,588 |
| Length | 110 |
| Max. P | 0.779842 |

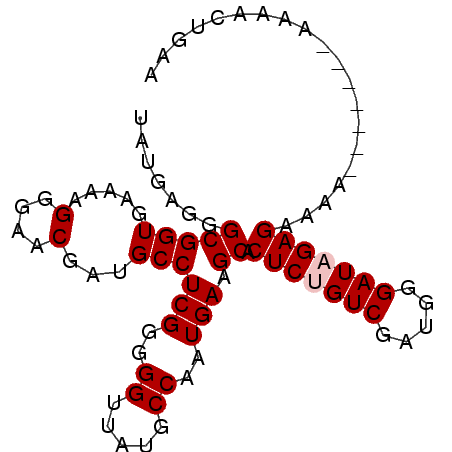
| Location | 14,597,478 – 14,597,569 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 92.62 |
| Mean single sequence MFE | -18.60 |
| Consensus MFE | -15.80 |
| Energy contribution | -16.20 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.779842 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14597478 91 + 20766785 UAUGAGGGCGGUGAAAAGGGAACGAUGCCUCGGGGGUUAUGCCAAUGAAGCACUCUGUCGAUGAGAUAGAGAAAAAAAAAAAAAAACUGAA ........((((.............(((.(((..((.....))..))).)))(((((((.....)))))))..............)))).. ( -21.10) >DroSec_CAF1 229625 83 + 1 UAUGAGGGCGGUGAAAAGGGAACGAUGCCUCGG-GGUUAUGCCAAUGAAGCACUCCGUCGAUGGGAUAGAGAAAA-------AAAACUGAA ......(((((.(....(....)..(((.(((.-((.....))..))).)))).)))))................-------......... ( -14.80) >DroSim_CAF1 232908 84 + 1 UAUGAGGGCGGUGAAAAGGGAACGAUGCCUCGGGGGUUAUGCCAAUGAAGCACUCUGUCGAUGGGAUAGAGAAAA-------AAAACUGAA ........((((.............(((.(((..((.....))..))).)))(((((((.....)))))))....-------...)))).. ( -20.50) >DroEre_CAF1 236985 83 + 1 UAUGAGGGCGGUGAAAAGGGAACGAGGCCUCGGGGGUUAUGCCAAUGAAGCUCUCCGUCGAUGGGAUAGAGAAAA-------AGAA-UGAA ......(((((.((...(....)...((.(((..((.....))..))).)))).)))))................-------....-.... ( -17.50) >DroYak_CAF1 245098 83 + 1 UAUGAGGGCGGUGAAAAGGGAACGAUGCCUCGGGGGUUAUGCCAAUGAAGCUCUCUGUCGAUGGGAUAGAGAAAA-------AGAA-UGAA .......((((((....(....)..))))(((..((.....))..))).))((((((((.....))))))))...-------....-.... ( -19.10) >consensus UAUGAGGGCGGUGAAAAGGGAACGAUGCCUCGGGGGUUAUGCCAAUGAAGCACUCUGUCGAUGGGAUAGAGAAAA_______AAAACUGAA .......(((((.....(....)...)))(((..((.....))..))).)).(((((((.....))))))).................... (-15.80 = -16.20 + 0.40)

| Location | 14,597,478 – 14,597,569 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 92.62 |
| Mean single sequence MFE | -11.76 |
| Consensus MFE | -10.74 |
| Energy contribution | -10.50 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.584023 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14597478 91 - 20766785 UUCAGUUUUUUUUUUUUUUUCUCUAUCUCAUCGACAGAGUGCUUCAUUGGCAUAACCCCCGAGGCAUCGUUCCCUUUUCACCGCCCUCAUA ....................((((.((.....)).))))((((((...((.....))...))))))......................... ( -11.50) >DroSec_CAF1 229625 83 - 1 UUCAGUUUU-------UUUUCUCUAUCCCAUCGACGGAGUGCUUCAUUGGCAUAACC-CCGAGGCAUCGUUCCCUUUUCACCGCCCUCAUA .........-------....(((..((.....))..)))((((((...((.....))-..))))))......................... ( -11.20) >DroSim_CAF1 232908 84 - 1 UUCAGUUUU-------UUUUCUCUAUCCCAUCGACAGAGUGCUUCAUUGGCAUAACCCCCGAGGCAUCGUUCCCUUUUCACCGCCCUCAUA .........-------....((((.((.....)).))))((((((...((.....))...))))))......................... ( -11.00) >DroEre_CAF1 236985 83 - 1 UUCA-UUCU-------UUUUCUCUAUCCCAUCGACGGAGAGCUUCAUUGGCAUAACCCCCGAGGCCUCGUUCCCUUUUCACCGCCCUCAUA ....-....-------...((((..((.....))..))))(((.....))).........((((...((............)).))))... ( -12.40) >DroYak_CAF1 245098 83 - 1 UUCA-UUCU-------UUUUCUCUAUCCCAUCGACAGAGAGCUUCAUUGGCAUAACCCCCGAGGCAUCGUUCCCUUUUCACCGCCCUCAUA ....-....-------...(((((.((.....)).)))))(((((...((.....))...))))).......................... ( -12.70) >consensus UUCAGUUUU_______UUUUCUCUAUCCCAUCGACAGAGUGCUUCAUUGGCAUAACCCCCGAGGCAUCGUUCCCUUUUCACCGCCCUCAUA ....................((((.((.....)).)))).(((((...((.....))...))))).......................... (-10.74 = -10.50 + -0.24)

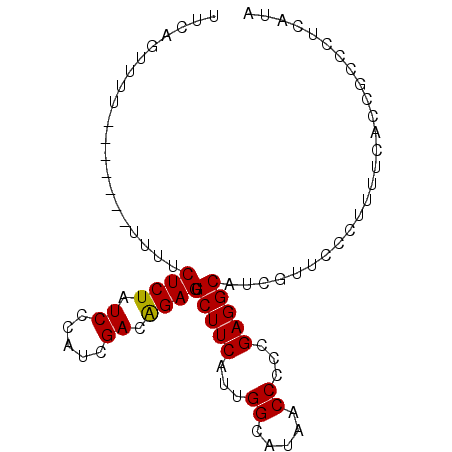
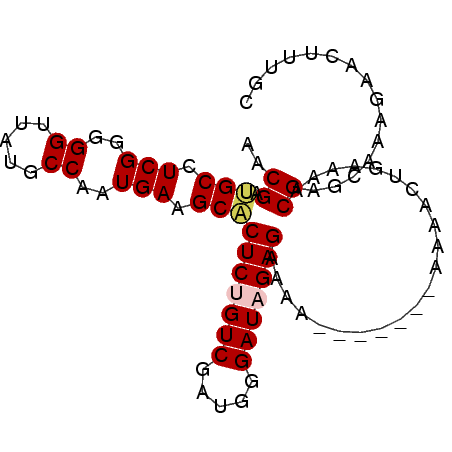
| Location | 14,597,498 – 14,597,588 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 84.36 |
| Mean single sequence MFE | -19.56 |
| Consensus MFE | -15.46 |
| Energy contribution | -15.82 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.710537 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14597498 90 + 20766785 AACGAUGCCUCGGGGGUUAUGCCAAUGAAGCACUCUGUCGAUGAGAUAGAGAAAAAAAAAAAAAAACUGAAAACGAGCAAAGAACUUUGC ..((.(((.(((..((.....))..))).)))(((((((.....)))))))......................)).(((((....))))) ( -22.30) >DroSec_CAF1 229645 69 + 1 AACGAUGCCUCGG-GGUUAUGCCAAUGAAGCACUCCGUCGAUGGGAUAGAGAAAA-------AAAACUGAAAGCGAG------------- .....(((.((((-..((.(((.......)))(((..((.....))..)))....-------))..))))..)))..------------- ( -12.00) >DroSim_CAF1 232928 83 + 1 AACGAUGCCUCGGGGGUUAUGCCAAUGAAGCACUCUGUCGAUGGGAUAGAGAAAA-------AAAACUGAAAGCGAGCAGAGAACUUUGC ..((.(((.(((..((.....))..))).)))(((((((.....)))))))....-------...........)).(((((....))))) ( -21.50) >DroEre_CAF1 237005 82 + 1 AACGAGGCCUCGGGGGUUAUGCCAAUGAAGCUCUCCGUCGAUGGGAUAGAGAAAA-------AGAA-UGAAAACGAGGCAAGAAUUUUGC ......((((((..((.....))........((((..((.....))..))))...-------....-......))))))........... ( -20.30) >DroYak_CAF1 245118 82 + 1 AACGAUGCCUCGGGGGUUAUGCCAAUGAAGCUCUCUGUCGAUGGGAUAGAGAAAA-------AGAA-UGAAAACGAGCAAAGAACUUUGC ..((..((.(((..((.....))..))).))((((((((.....))))))))...-------....-......)).(((((....))))) ( -21.70) >consensus AACGAUGCCUCGGGGGUUAUGCCAAUGAAGCACUCUGUCGAUGGGAUAGAGAAAA_______AAAACUGAAAACGAGCAAAGAACUUUGC ..((.(((.(((..((.....))..))).)))(((((((.....)))))))......................))............... (-15.46 = -15.82 + 0.36)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:19:39 2006