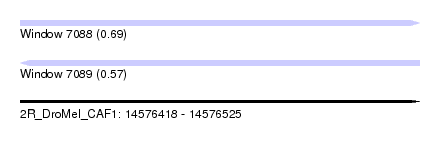
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,576,418 – 14,576,525 |
| Length | 107 |
| Max. P | 0.690247 |

| Location | 14,576,418 – 14,576,525 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 83.28 |
| Mean single sequence MFE | -33.62 |
| Consensus MFE | -22.84 |
| Energy contribution | -24.28 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.690247 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
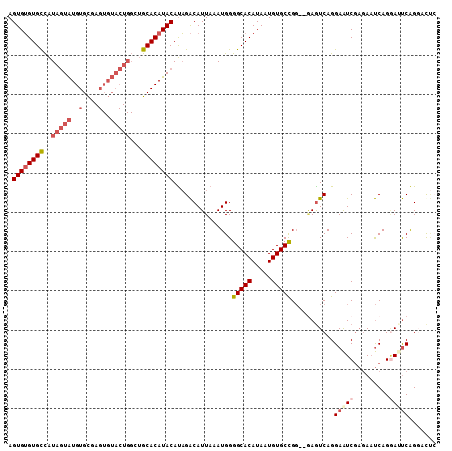
>2R_DroMel_CAF1 14576418 107 + 20766785 AGUGUGUGCCAUAGUAUGUGCAAGUGUACUGGCUGCACAUACAUAGACAUUAAAUGGGGCACAUAAUGUGCCGG--GAGUCAGGAAUCGAGAAUCAGAAUUCAGGCUUC .((((((((..(((((..(....)..)))))...))))))))...............(((((.....))))).(--(((((.(((.((........)).))).)))))) ( -35.10) >DroSec_CAF1 208780 107 + 1 AGUGUGUGCCAUAGUAUGUGCGAGUGUACUGGCUGCACAUACAUAGACAUUAAAUGGGGCACAUAAUGUGCUGG--UAGUCAGGAAUCGAGAAUCAGGAUUCAGGCUUC ..((((((((...((((((((.(((......))))))))))).....(((...))).))))))))....(((.(--.((((..((........))..))))).)))... ( -32.40) >DroSim_CAF1 210247 107 + 1 AGUGUGUGCCAUAGUAUGUGCGAGUGUACUGGCUGCACAUACAUAGACAUUAAAUGGGGCACAUAAUGUGCUGG--GAGUCAGGAAUCGAGAAUCAGGAUUCAGGCUUC ..((((((((...((((((((.(((......))))))))))).....(((...))).))))))))....(((..--(((((..((........))..))))).)))... ( -36.10) >DroEre_CAF1 215551 105 + 1 AGUGUGUGCCAUAGUAUGUGCUAGUGUACUGGGUGCACAUACAUAGACAUUAAAUGGGGCACAUAAUGUGCCGG--GAGUCAGGAAUCGAGAAUCAGGAAUCAGGAC-- .(((((..((.(((((..(....)..)))))))..))))).................(((((.....)))))..--..(((..((.((.........)).))..)))-- ( -31.40) >DroYak_CAF1 222713 109 + 1 AGUGUGUGCCAUAGUAUGUGCUAGUGUACUGGGUGCACAUACAUAGACAUUAAAUGGGGCACAUAAUGUGCCGGAGAAAUCAGGAAUCGAGAAUCAGGAAUCAGUACUA .(((((..((.(((((..(....)..)))))))..)))))...(((((.........(((((.....))))).((....))......................)).))) ( -30.50) >DroAna_CAF1 247302 95 + 1 AGUGGGUGUUGUACUGUGUGUG------------GCACAUACACUGGCAUUAAAUGGGGCACAUAAUGUGCUGC--GAGUCAGGAGUCACUACCCAGUACCCGGCACCC ...(((((((((((((.(((((------------((.......(((((......((.(((((.....))))).)--).)))))..))))).)).))))))..))))))) ( -36.20) >consensus AGUGUGUGCCAUAGUAUGUGCGAGUGUACUGGCUGCACAUACAUAGACAUUAAAUGGGGCACAUAAUGUGCCGG__GAGUCAGGAAUCGAGAAUCAGGAUUCAGGACUC .((((((((..(((((..(....)..)))))...))))))))...............(((((.....)))))...........(((((.........)))))....... (-22.84 = -24.28 + 1.45)

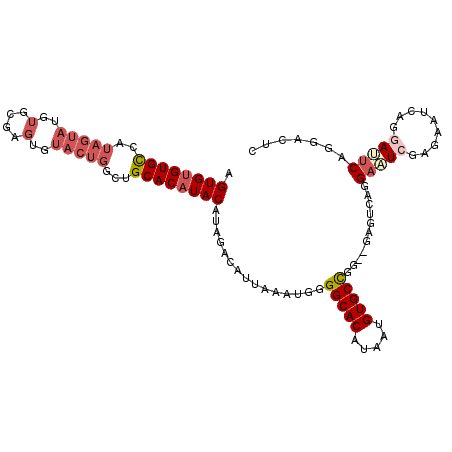
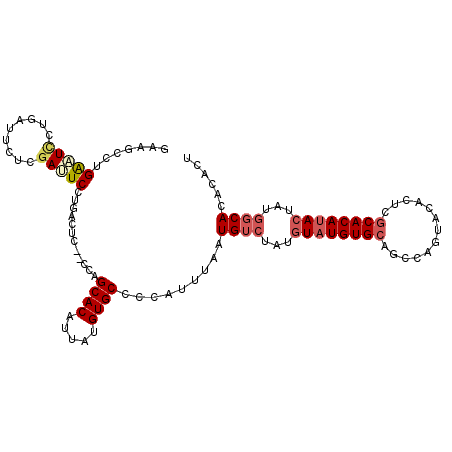
| Location | 14,576,418 – 14,576,525 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 83.28 |
| Mean single sequence MFE | -26.12 |
| Consensus MFE | -17.03 |
| Energy contribution | -17.95 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.573814 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14576418 107 - 20766785 GAAGCCUGAAUUCUGAUUCUCGAUUCCUGACUC--CCGGCACAUUAUGUGCCCCAUUUAAUGUCUAUGUAUGUGCAGCCAGUACACUUGCACAUACUAUGGCACACACU (((....((((....))))....))).......--..(((((.....)))))........((((...((((((((((.........))))))))))...))))...... ( -27.20) >DroSec_CAF1 208780 107 - 1 GAAGCCUGAAUCCUGAUUCUCGAUUCCUGACUA--CCAGCACAUUAUGUGCCCCAUUUAAUGUCUAUGUAUGUGCAGCCAGUACACUCGCACAUACUAUGGCACACACU (((....((((....))))....))).......--...((((.....)))).........((((...((((((((((........)).))))))))...))))...... ( -23.90) >DroSim_CAF1 210247 107 - 1 GAAGCCUGAAUCCUGAUUCUCGAUUCCUGACUC--CCAGCACAUUAUGUGCCCCAUUUAAUGUCUAUGUAUGUGCAGCCAGUACACUCGCACAUACUAUGGCACACACU (((....((((....))))....))).......--...((((.....)))).........((((...((((((((((........)).))))))))...))))...... ( -23.90) >DroEre_CAF1 215551 105 - 1 --GUCCUGAUUCCUGAUUCUCGAUUCCUGACUC--CCGGCACAUUAUGUGCCCCAUUUAAUGUCUAUGUAUGUGCACCCAGUACACUAGCACAUACUAUGGCACACACU --(((..((.((.........)).))..)))..--..(((((.....)))))........((((...((((((((.............))))))))...))))...... ( -25.72) >DroYak_CAF1 222713 109 - 1 UAGUACUGAUUCCUGAUUCUCGAUUCCUGAUUUCUCCGGCACAUUAUGUGCCCCAUUUAAUGUCUAUGUAUGUGCACCCAGUACACUAGCACAUACUAUGGCACACACU .....................................(((((.....)))))........((((...((((((((.............))))))))...))))...... ( -23.22) >DroAna_CAF1 247302 95 - 1 GGGUGCCGGGUACUGGGUAGUGACUCCUGACUC--GCAGCACAUUAUGUGCCCCAUUUAAUGCCAGUGUAUGUGC------------CACACACAGUACAACACCCACU (((((....((((((((((((((........))--)).((((.....)))).........)))).((((......------------.)))).))))))..)))))... ( -32.80) >consensus GAAGCCUGAAUCCUGAUUCUCGAUUCCUGACUC__CCAGCACAUUAUGUGCCCCAUUUAAUGUCUAUGUAUGUGCAGCCAGUACACUCGCACAUACUAUGGCACACACU .......(((((.........)))))............((((.....)))).........((((...((((((((.............))))))))...))))...... (-17.03 = -17.95 + 0.92)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:19:34 2006