
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
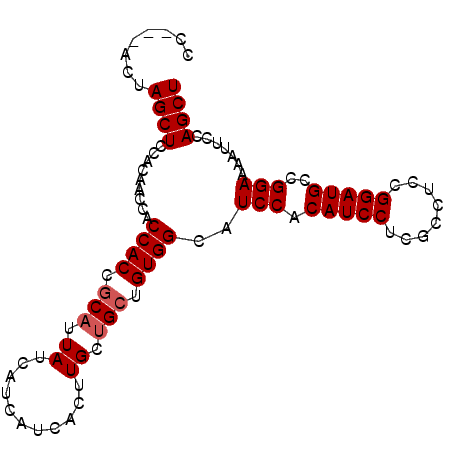
| Location | 14,574,837 – 14,574,928 |
| Length | 91 |
| Max. P | 0.999604 |

| Location | 14,574,837 – 14,574,928 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 92.62 |
| Mean single sequence MFE | -19.85 |
| Consensus MFE | -17.84 |
| Energy contribution | -18.24 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.90 |
| SVM decision value | 3.77 |
| SVM RNA-class probability | 0.999604 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
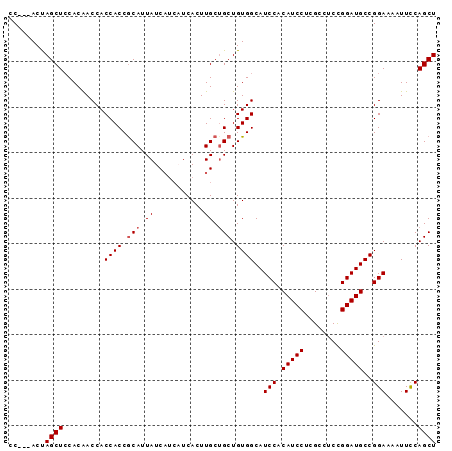
>2R_DroMel_CAF1 14574837 91 + 20766785 CC---ACUAGCUCCACAACCACCACCGCAUUAUCAUCAUCACUUGCUGCUGUGGCAUCCACAUCCUCGCCUCCGGAUGCCGGAAAAUUUCAGCU ..---...((((.........((((.(((.((...........)).))).))))..(((.(((((........)))))..))).......)))) ( -19.30) >DroSec_CAF1 207233 88 + 1 CC---ACUAGCUACACAACCACCACCGCAUUAUC---AUCACUUGCUGCUGUGGCAUCCACAUCCUCGCCUCUGGAUGCCGGAAAAUUCCAGCU ..---...((((.........((((.(((.((..---......)).))).))))..(((.(((((........)))))..))).......)))) ( -21.50) >DroSim_CAF1 208689 91 + 1 CC---ACUAGCUCCACAACCACCACCGCAUUAUCAUCAUCACUUGCUGCUGUGGCAUCCACAUCCUCGCCUCCGGAUGCCGGAAAAUUCCAGCU ..---...((((.........((((.(((.((...........)).))).))))..(((.(((((........)))))..))).......)))) ( -19.30) >DroEre_CAF1 213987 91 + 1 CC---ACCAGCUCCACGACCACCACCACAUUAUCAUCCUCACUUGCUGCUGUGGCAUCCACAUCCCCGCCGCCGGAUGCCGGAAAAUUCCAGCU .(---(.((((.................................)))).)).(((((((..............)))))))(((....))).... ( -18.05) >DroYak_CAF1 221149 94 + 1 CCAGCACCAGCUCCACAACCACCACCGCAUUAUCAUCCUCACUUGCAGCUGUGGCAUCCACAUCCUCGCCUCCGGAUGCCGGAAAAUUCCAGCU ..(((..(((((..............(((..............)))))))).(((((((..............)))))))(((....))).))) ( -21.11) >consensus CC___ACUAGCUCCACAACCACCACCGCAUUAUCAUCAUCACUUGCUGCUGUGGCAUCCACAUCCUCGCCUCCGGAUGCCGGAAAAUUCCAGCU ........((((.........((((.(((.((...........)).))).))))..(((.(((((........)))))..))).......)))) (-17.84 = -18.24 + 0.40)

| Location | 14,574,837 – 14,574,928 |
|---|---|
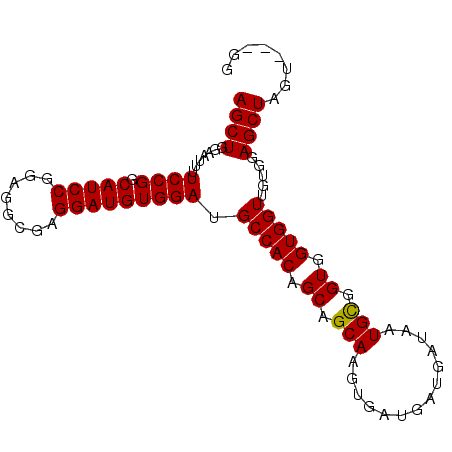
| Length | 91 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 92.62 |
| Mean single sequence MFE | -28.84 |
| Consensus MFE | -24.78 |
| Energy contribution | -24.62 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.758744 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
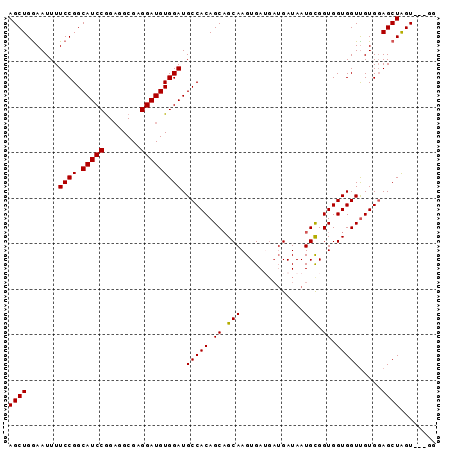
>2R_DroMel_CAF1 14574837 91 - 20766785 AGCUGAAAUUUUCCGGCAUCCGGAGGCGAGGAUGUGGAUGCCACAGCAGCAAGUGAUGAUGAUAAUGCGGUGGUGGUUGUGGAGCUAGU---GG ((((.......((((.(((((........)))))))))..(((((((.((.(.((((.......)).)).).)).)))))))))))...---.. ( -26.80) >DroSec_CAF1 207233 88 - 1 AGCUGGAAUUUUCCGGCAUCCAGAGGCGAGGAUGUGGAUGCCACAGCAGCAAGUGAU---GAUAAUGCGGUGGUGGUUGUGUAGCUAGU---GG .((((((....))))))..(((..(((....(((..(..(((((.(((.((.....)---)....))).)))))..)..))).)))..)---)) ( -28.30) >DroSim_CAF1 208689 91 - 1 AGCUGGAAUUUUCCGGCAUCCGGAGGCGAGGAUGUGGAUGCCACAGCAGCAAGUGAUGAUGAUAAUGCGGUGGUGGUUGUGGAGCUAGU---GG .(((((.....((((.(((((........)))))))))..(((((((.((.(.((((.......)).)).).)).)))))))..)))))---.. ( -27.90) >DroEre_CAF1 213987 91 - 1 AGCUGGAAUUUUCCGGCAUCCGGCGGCGGGGAUGUGGAUGCCACAGCAGCAAGUGAGGAUGAUAAUGUGGUGGUGGUCGUGGAGCUGGU---GG .((((((....))))))..(((((..(.(.(((.....(((((((....((........))....)))))))...))).).).))))).---.. ( -29.80) >DroYak_CAF1 221149 94 - 1 AGCUGGAAUUUUCCGGCAUCCGGAGGCGAGGAUGUGGAUGCCACAGCUGCAAGUGAGGAUGAUAAUGCGGUGGUGGUUGUGGAGCUGGUGCUGG .((((((....))))))(((((.(........).))))).(((((((..(......(.((....)).)....)..)))))))(((....))).. ( -31.40) >consensus AGCUGGAAUUUUCCGGCAUCCGGAGGCGAGGAUGUGGAUGCCACAGCAGCAAGUGAUGAUGAUAAUGCGGUGGUGGUUGUGGAGCUAGU___GG ((((.......((((.(((((........))))))))).(((((.((.(((..............))).)).))))).....))))........ (-24.78 = -24.62 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:19:32 2006