
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,554,793 – 14,554,883 |
| Length | 90 |
| Max. P | 0.879756 |

| Location | 14,554,793 – 14,554,883 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 77.51 |
| Mean single sequence MFE | -20.82 |
| Consensus MFE | -15.38 |
| Energy contribution | -16.22 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.879756 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14554793 90 + 20766785 CUGGACCUGCACAUUUCUGGCCAAAAUGCCACGAAUUGUAUGCCACGGGUCCGGCUCCACUUCCCCACACUGCCAACGGUCCCACAAAAA ..(((((((((((.(((((((......)))).))).)))..))...))))))(((................)))................ ( -23.99) >DroSec_CAF1 187380 85 + 1 CUGGACCUGCACAUUUCUGGCCAAAAUGCCCCGAAUUGUAUGCCACGGGUCCGGCUCCUCUCGCCUC-----CUCCUCGCCCCACAAAAA ..(((((((((((.(((.(((......)))..))).)))..))...))))))(((.......)))..-----.................. ( -21.40) >DroSim_CAF1 188663 90 + 1 CUGGACCUGCACAUUUCUGGCCAAAAUGCCCCGAAUUGUAUGCCACGGGUCCGGCUCCUCUCGCCCGCAAUUCCCACGGCCCCACAAAAA (((((((((((((.(((.(((......)))..))).)))..))...))))))))........(((............))).......... ( -21.90) >DroEre_CAF1 193587 71 + 1 CUGGACCUGCACAUUUCUGGCCAAAAUGCCUCGAAUUGUAUGCCAUGGGCCCUGCUGCUCA--------------C----CCCACAA-AA .(((.(.((((.((((..(((......)))..)))))))).))))(((((......)))))--------------.----.......-.. ( -15.80) >DroYak_CAF1 200541 76 + 1 CUGGACCUGCACAUUUCUGGCCCAAAUGCCCCGAAUUGUAUGCCAUAGGGCCUCUCCCUCC--------------CCGGGCCCACAAAAA .(((.(.((((.((((..(((......)))..)))))))).))))..((((((........--------------..))))))....... ( -21.00) >consensus CUGGACCUGCACAUUUCUGGCCAAAAUGCCCCGAAUUGUAUGCCACGGGUCCGGCUCCUCU__CC_______C__CCGGCCCCACAAAAA (((((((((((((.(((.(((......)))..))).)))..))...)))))))).................................... (-15.38 = -16.22 + 0.84)

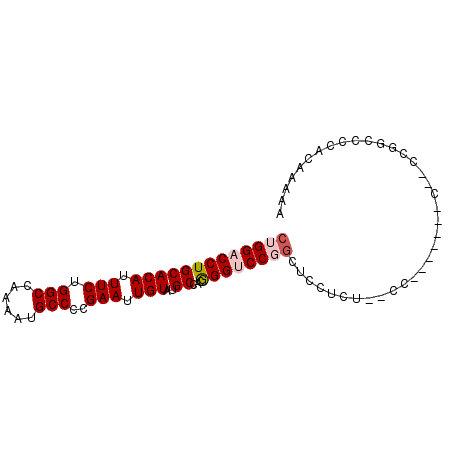
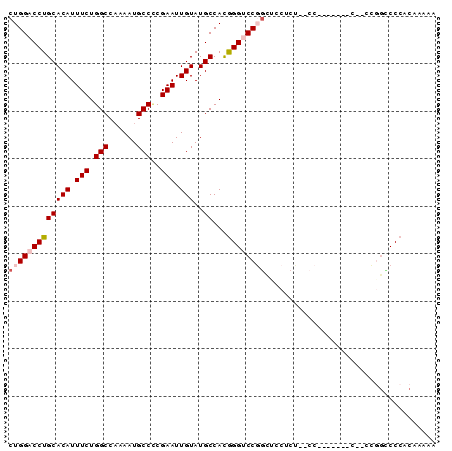
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:19:24 2006