| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,532,948 – 14,533,056 |
| Length | 108 |
| Max. P | 0.500000 |

| Location | 14,532,948 – 14,533,056 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 81.58 |
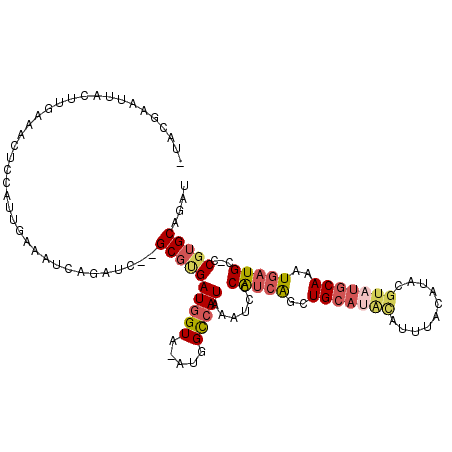
| Mean single sequence MFE | -26.25 |
| Consensus MFE | -15.80 |
| Energy contribution | -16.83 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.60 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
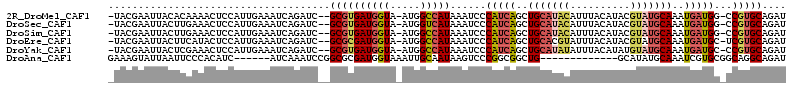
>2R_DroMel_CAF1 14532948 108 + 20766785 -UACGAAUUACACAAAACUCCAUUGAAAUCAGAUC--GCGUGAUGGUA-AUGGCCAUAAAUCCCAUCAGCUGCAUACAUUUACAUACGUAUGCAAAUGAUGG-CCGUGCAGAU -.........(((......((((((..((((....--...))))..))-)))).........((((((..(((((((..........)))))))..))))))-..)))..... ( -28.40) >DroSec_CAF1 165243 108 + 1 -UACGAAUUACUUGAAACUCCAUUGAAAUCAGAUC--GCGUGAUGGUA-AUGGUCAUAAAUCCCAUCAGCUGCAUACAUUUACAUACGUAUGCAAAUGAUGG-CCGUGCAGAU -((((..............((((((..((((....--...))))..))-)))).........((((((..(((((((..........)))))))..))))))-.))))..... ( -28.40) >DroSim_CAF1 166444 108 + 1 -UACGAAUUACUUGAAACUCCAUUGAAAUCAGAUC--GCGUGAUGGUA-AUGGCCAUAAAUCCCAUCAGCUGCAUACAUUUACAUACGUAUGCAAAUGAUGG-CCGUGCAGAU -((((..............((((((..((((....--...))))..))-)))).........((((((..(((((((..........)))))))..))))))-.))))..... ( -28.20) >DroEre_CAF1 171577 108 + 1 -UACGAAUUACUUCAUACUCCAUUGAAAUCAGAUC--GCGCGAUGGUA-AUGGCCAUAAAUCCCAUCAGCUGCACGUAUUUACAUACGUAUGCAAAUGAUGC-UCGUGCAGAU -..........((((........))))........--((((((((((.-...)))).......(((((..(((((((((....))))))..)))..))))).-)))))).... ( -27.20) >DroYak_CAF1 169779 108 + 1 -UACGAAUUACUCGAAACUCCAUUGAAAUCAGAUC--GCGUGAUGGUA-AUGGCCAUAAAUCCCAUCAGCUGCAUAUAUUUACAUAUGUAUGCAAAUGAUGC-CCGUGCAGAU -((((..............((((((..((((....--...))))..))-))))..........(((((..(((((((((......)))))))))..))))).-.))))..... ( -26.60) >DroAna_CAF1 185128 94 + 1 GAAAGUAUUAAUUCCCACAUC------AUCAAAUCCGGCGCGAUGGUAAAUUGCAAUAAGUCCCGGCGGCUG-------------GCAUAUGCAAAUCGUGCGGCAGGCAGAU .....................------.......(((.((((((.(((...(((....((((.....)))).-------------)))..)))..)))))))))......... ( -18.70) >consensus _UACGAAUUACUUGAAACUCCAUUGAAAUCAGAUC__GCGUGAUGGUA_AUGGCCAUAAAUCCCAUCAGCUGCAUACAUUUACAUACGUAUGCAAAUGAUGC_CCGUGCAGAU .....................................((((((((((.....)))))......(((((..(((((((..........)))))))..)))))...))))).... (-15.80 = -16.83 + 1.03)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:19:16 2006