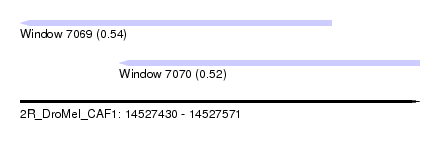
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,527,430 – 14,527,571 |
| Length | 141 |
| Max. P | 0.535910 |

| Location | 14,527,430 – 14,527,540 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 76.64 |
| Mean single sequence MFE | -28.94 |
| Consensus MFE | -13.52 |
| Energy contribution | -12.86 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.535910 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
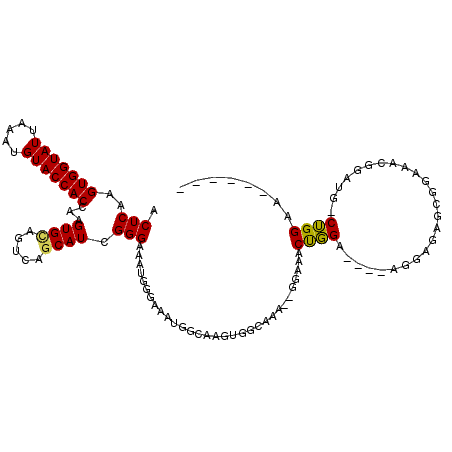
>2R_DroMel_CAF1 14527430 110 - 20766785 ACUCAAGUGGUAUUAAAUGUACCACCAAGUGCAGUCGGCAUCGGGAAAUGGGAAAUGGCAAGUGGCAAA--GGAAACUGGA----AGGAGAGCGGAAACGAAUG-CUGGAAGGCGGC .((((..((((((.....))))))((..((((.....))))..))...)))).....((....((((.(--(....))...----.......((....))..))-)).....))... ( -28.10) >DroPse_CAF1 156669 111 - 1 ACUCAAGUGGUAUUAAAUGUACCACCAAGUGUAGUCAGCAUCGGGAAGUGGCAAGUGCCAGUUGAAGAGGGGGAUACCCGAGAACAGGAGACGGGCAAUGGACUACGGGCA------ .(((..(((((((.....))))))).....((((((.((.(((..((.((((....)))).)).....(((.....)))............)))))....)))))))))..------ ( -35.30) >DroSim_CAF1 160830 107 - 1 ACUCAAGUGGUAUUAAAUGUACCACCAAGUGCAGUCGACAUCGGGAAAUGGGAAAUGGCAAGUGGCAAA--GGAAACUGGA----AGGAGAGCGGAAACGGAUG-CUGGAAGG---U .(((..(((((((.....)))))))((..(((..((..(((......))).))....)))..))....(--(....))...----..)))..((....))....-........---. ( -24.60) >DroYak_CAF1 163820 107 - 1 ACUCAAGUGGUAUUAAAUGUACCACCAAGUGCAGUCGGCAUCGGGAAAUGGGAAAUGGCAAGUGGCAAA--GGAAACUGGA----AGGAGAGCGGAAACGGAUG-CUGGAAGU---C (((...(((((((.....)))))))..)))((..((((((((.............((.(....).)).(--(....))...----.......((....))))))-))))..))---. ( -28.50) >DroAna_CAF1 177690 87 - 1 ACUCAAGUGGUAUUAAAUGUACCACCAAGUGCAGUCAGCAUCGGGAA-------AUGGCAAGUGGCAAC--UGAAACUGGA----AA-----CGGAAA-----A-CUGGGA------ .((((((((((((.....))))).(((..(((..((........)).-------...)))..)))..))--)....(((..----..-----)))...-----.-.)))).------ ( -22.00) >DroPer_CAF1 158860 111 - 1 ACUCAAGUGGUAUUAAAUGUACCACCAAGUGUAGUCAGCAUCGGGAAGUGGCAAGUGCCAGUUGAAGAGGGGGAUACCCGAGAACAGGAGACAGGCAAUGGACUACGGGCA------ .(((..(((((((.....))))))).....((((((.((.(((((((.((((....)))).)).............))))).....(....)..))....)))))))))..------ ( -35.11) >consensus ACUCAAGUGGUAUUAAAUGUACCACCAAGUGCAGUCAGCAUCGGGAAAUGGGAAAUGGCAAGUGGCAAA__GGAAACUGGA____AGGAGAGCGGAAACGGAUG_CUGGAA______ .(((..(((((((.....)))))))...((((.....)))).)))...............................((((.........................))))........ (-13.52 = -12.86 + -0.66)


| Location | 14,527,465 – 14,527,571 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 82.00 |
| Mean single sequence MFE | -32.55 |
| Consensus MFE | -18.84 |
| Energy contribution | -18.87 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.58 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.519631 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
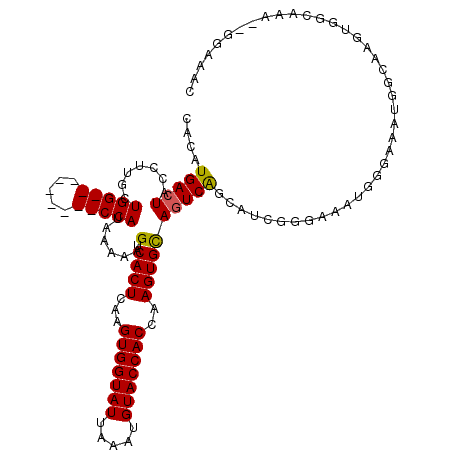
>2R_DroMel_CAF1 14527465 106 - 20766785 CACAUGACUACGUUGCUGG---------CCAUAAAAAGUCACUCAAGUGGUAUUAAAUGUACCACCAAGUGCAGUCGGCAUCGGGAAAUGGGAAAUGGCAAGUGGCAAA--GGAAAC ...........((..((.(---------((((.....((..(((..(((((((.....)))))))...((((.....)))).)))..)).....))))).))..))...--...... ( -28.50) >DroPse_CAF1 156703 117 - 1 UGUAUGACUGCCUUGCUGGCCACGGUGGCCAUAAAAAGUCACUCAAGUGGUAUUAAAUGUACCACCAAGUGUAGUCAGCAUCGGGAAGUGGCAAGUGCCAGUUGAAGAGGGGGAUAC .......((.(((((((((((((((((((........))))))...(((((((.....)))))))...)))..))))))......((.((((....)))).))...)))).)).... ( -40.70) >DroSec_CAF1 159751 106 - 1 CACAUGACUACGUUGCUGG---------CCAUAAAAAGUCACUCAAGUGGUAUUAAAUGUACCACCAAGUGCAGUCGGCAGCGGGAAAUGGGAAAUGGCAAGUGGCAAA--GGAAAC ..(((..((.(((((((((---------(..........((((...(((((((.....)))))))..))))..))))))))))))..))).....((.(....).))..--...... ( -28.60) >DroSim_CAF1 160862 106 - 1 CACAUGACUACGUUGCUGG---------CCAUAAAAAGUCACUCAAGUGGUAUUAAAUGUACCACCAAGUGCAGUCGACAUCGGGAAAUGGGAAAUGGCAAGUGGCAAA--GGAAAC ...........((..((.(---------((((.....(.((((...(((((((.....)))))))..)))))..(((....)))..........))))).))..))...--...... ( -28.70) >DroAna_CAF1 177709 99 - 1 CACAUGAGUGCCCGCCUGG---------CCAUAAAAAGUCACUCAAGUGGUAUUAAAUGUACCACCAAGUGCAGUCAGCAUCGGGAA-------AUGGCAAGUGGCAAC--UGAAAC ....(.(((..((((...(---------((((...............((((((.....))))))((..((((.....))))..))..-------)))))..))))..))--).)... ( -28.10) >DroPer_CAF1 158894 117 - 1 UGUAUGACUGCCUUGCUGGCCACGGUGGCCAUAAAAAGUCACUCAAGUGGUAUUAAAUGUACCACCAAGUGUAGUCAGCAUCGGGAAGUGGCAAGUGCCAGUUGAAGAGGGGGAUAC .......((.(((((((((((((((((((........))))))...(((((((.....)))))))...)))..))))))......((.((((....)))).))...)))).)).... ( -40.70) >consensus CACAUGACUACCUUGCUGG_________CCAUAAAAAGUCACUCAAGUGGUAUUAAAUGUACCACCAAGUGCAGUCAGCAUCGGGAAAUGGGAAAUGGCAAGUGGCAAA__GGAAAC ....(((((.......(((((.....)))))......(.((((...(((((((.....)))))))..))))))))))........................................ (-18.84 = -18.87 + 0.03)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:19:15 2006