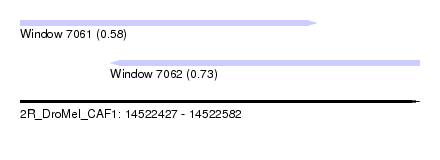
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,522,427 – 14,522,582 |
| Length | 155 |
| Max. P | 0.730944 |

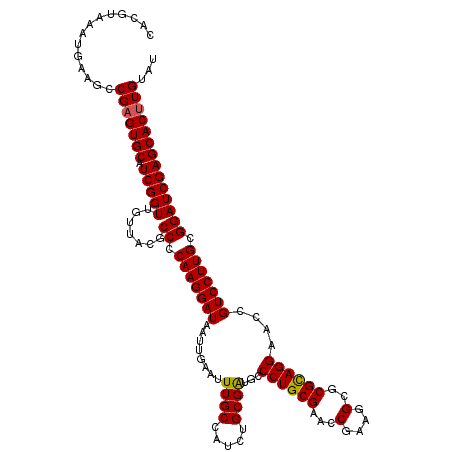
| Location | 14,522,427 – 14,522,542 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.11 |
| Mean single sequence MFE | -33.75 |
| Consensus MFE | -26.28 |
| Energy contribution | -26.33 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.583210 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14522427 115 + 20766785 UGUUCUU-----ACUUUUCGCUUGCAGAUACAAUCAUCGCAUACAAGUGCUCGAUCCGCAAGGACGAUUCCUGCGCGCCUUCGGUUCGCAGGGCACCGCAGAUGGCAAAUUCAAUUAUCC .......-----.......((((((.(((......)))........(((((((.(((....))))))..((((((.(((...))).)))))))))).)))...))).............. ( -34.20) >DroVir_CAF1 198307 115 + 1 UCAAC-----UGUCUACUAUGUUGCAGAUACAAUCAUCGCAUACAAGUGCUCGAUCCACAAGGACGGUUCCUGCGCGCCUUCGGCUCGCAGGGCAUCGCAGAUGGCAAAUUCAAUUAUCC ....(-----((((((((.((((((.(((......)))))).))))))((..(.(((....))))((((((((((.(((...))).))))))).)))))))))))............... ( -32.40) >DroGri_CAF1 176675 116 + 1 UCAA-AU---UGUCUACUAUGUUGCAGAUAUAAUCAUCGCAUACAAGUGCUCGAUCCACAAGGACGCUUCCUACGUGCCUUCGGUUCGCAGGGCAUCGCGGAUGGCAAAUUCAAUUAUCC ....-.(---((((((((.((((((.(((......)))))).)))))))....((((.(.((((....))))..(((((((((...)).))))))).).)))))))))............ ( -26.80) >DroWil_CAF1 234637 120 + 1 UGUUAUUUGCUCUCUGUUUCUUACCAGAUACAAUCAUCGCAUACAAGUGCUUGAUCCGCAAGGACGUUUCCUGCGCGCCUUCGGUUCGCAGGGCACCGCAGAUGGCAAAUUCAAUUAUCC ((((((((((..((((........))))..................((((....(((....))).....((((((.(((...))).)))))))))).))))))))))............. ( -38.60) >DroMoj_CAF1 246426 117 + 1 UCACCUU---UGUUUGCCACAUUGCAGAUACAAUCAUCGCAUUCAGGUGCUCGAUCCGCAAGGACGUUUCCUGCGCGCCUUCGGUUCGCAGGGCAUCGCAGAUGGCAAAUUCAAUUAUCC .......---.((((((((..((((.(((......))).......(((((.((.(((....)))))...((((((.(((...))).))))))))))))))).)))))))).......... ( -41.50) >DroPer_CAF1 153633 113 + 1 --UUACU-----ACUCUUUACUUCCAGAUAUAAUCACCGCAUACAAGUGCUCGAUCCGCAAGGACGGUUCCUGCGCGCAUUUGGUUCGCAGGGGACCGCAGAUGGCAAAUUCAAUUAUCC --.....-----...........(((............((((....))))((..(((....)))(((((((((((.((.....)).)))).)))))))..)))))............... ( -29.00) >consensus UCAUCUU___UGUCUACUACCUUGCAGAUACAAUCAUCGCAUACAAGUGCUCGAUCCGCAAGGACGGUUCCUGCGCGCCUUCGGUUCGCAGGGCACCGCAGAUGGCAAAUUCAAUUAUCC ..........................((.....(((((((((....)))).((.(((....)))))..(((((((.((.....)).))))))).......))))).....))........ (-26.28 = -26.33 + 0.06)

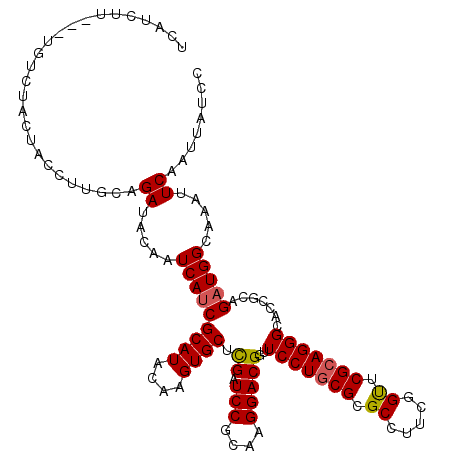
| Location | 14,522,462 – 14,522,582 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.06 |
| Mean single sequence MFE | -39.14 |
| Consensus MFE | -36.88 |
| Energy contribution | -36.66 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.00 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.730944 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14522462 120 - 20766785 CACGUAAAUGAAGCCGAGUGCAUCGGUUGUUACGCCCCAAGGAUAAUUGAAUUUGCCAUCUGCGGUGCCCUGCGAACCGAAGGCGCGCAGGAAUCGUCCUUGCGGAUCGAGCACUUGUAU ..............(((((((.(((((.......((.((((((..(((......(((......)))..((((((..((...))..)))))))))..)))))).))))))))))))))... ( -40.71) >DroVir_CAF1 198342 120 - 1 AACGUAAAUGAAGCCGAGUGCAUCGGUUGUUACGCCCCAAGGAUAAUUGAAUUUGCCAUCUGCGAUGCCCUGCGAGCCGAAGGCGCGCAGGAACCGUCCUUGUGGAUCGAGCACUUGUAU ..............(((((((.(((((.......((.(((((((........((((.....))))...((((((.(((...))).))))))....))))))).))))))))))))))... ( -42.61) >DroPse_CAF1 151459 120 - 1 CACGUAAAUGAAGCCGAGUGCAUCGGUUGUUACUCCCCAAGGAUAAUUGAAUUUGCCAUCUGCGGUCCCCUGCGAACCAAAUGCGCGCAGGAACCGUCCUUGCGGAUCGAGCACUUGUAU ..............(((((((.((((.......(((.(((((((..........((.....))(((..((((((..(.....)..)))))).)))))))))).))))))))))))))... ( -39.61) >DroGri_CAF1 176711 120 - 1 AACGUAAAUGAAGCCGAGUGCAUCGGUUGUUACGCCCCAAGGAUAAUUGAAUUUGCCAUCCGCGAUGCCCUGCGAACCGAAGGCACGUAGGAAGCGUCCUUGUGGAUCGAGCACUUGUAU ..(((((....((((((.....)))))).)))))...(((((((......)))(((((((((((((((((((((..((...))..))))))..)))))...)))))).).)))))))... ( -37.10) >DroMoj_CAF1 246463 120 - 1 UACGUAAAUGAAGCCGAGUGCAUCGGUUGUAACGCCCCAAGGAUAAUUGAAUUUGCCAUCUGCGAUGCCCUGCGAACCGAAGGCGCGCAGGAAACGUCCUUGCGGAUCGAGCACCUGAAU ..............((.((((.(((((.......((.(((((((........((((.....))))...((((((..((...))..))))))....))))))).))))))))))).))... ( -35.21) >DroPer_CAF1 153666 120 - 1 CACGUAAAUGAAGCCGAGUGCAUCGGUUGUUACUCCCCAAGGAUAAUUGAAUUUGCCAUCUGCGGUCCCCUGCGAACCAAAUGCGCGCAGGAACCGUCCUUGCGGAUCGAGCACUUGUAU ..............(((((((.((((.......(((.(((((((..........((.....))(((..((((((..(.....)..)))))).)))))))))).))))))))))))))... ( -39.61) >consensus CACGUAAAUGAAGCCGAGUGCAUCGGUUGUUACGCCCCAAGGAUAAUUGAAUUUGCCAUCUGCGAUGCCCUGCGAACCGAAGGCGCGCAGGAACCGUCCUUGCGGAUCGAGCACUUGUAU ..............(((((((.(((((.......((.(((((((........((((.....))))...((((((...(....)..))))))....))))))).))))))))))))))... (-36.88 = -36.66 + -0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:19:08 2006