| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,522,039 – 14,522,140 |
| Length | 101 |
| Max. P | 0.733836 |

| Location | 14,522,039 – 14,522,140 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 82.01 |
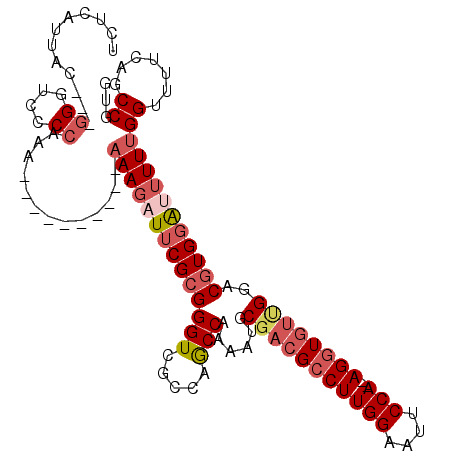
| Mean single sequence MFE | -37.08 |
| Consensus MFE | -22.99 |
| Energy contribution | -24.43 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.733836 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
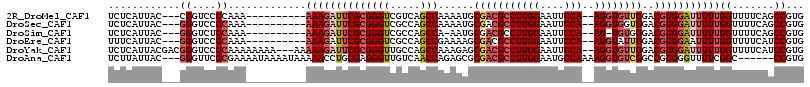
>2R_DroMel_CAF1 14522039 101 - 20766785 UCUCAUUAC---CGGUCCCCAAA----------AAAGAUUCGCGGGUCGUCAGCCAAAAUGCGACGCCUUGGAAUUCCA--AGGUGUUGGACGUGGAUUUUUGGUUUUCAGCCGUG .........---((((.......----------((((((((((((((.....)))......(((((((((((....)))--))))))))..)))))))))))........)))).. ( -37.06) >DroSec_CAF1 154376 101 - 1 UCUCAUUAC---GGGUCCCCAAA----------AAAGAUUCGCGGGUCGCCAGCCAAAAUGCGACGCCUUGGAAUUCCA--AGGUGGUGGACGUGGAUUUUUGGUUUUCAGCCGUG ......(((---((.(.......----------(((((((((((..(((((.((......))...(((((((....)))--))))))))).))))))))))).......).))))) ( -36.84) >DroSim_CAF1 155502 99 - 1 UCUCAUUAC---GGGUCUCCAAA----------AAAGAUUCGCGGGUCGCCAGCCA-AAUGCGACGCCUUGGAAUUCCA--AG-UGUGGGACGUGGAUUUUUGGUUUUCAGCCGUG ......(((---((.(..(((((----------...((((((((.(((((......-...)))))(((((((....)))--))-.))....))))))))))))).....).))))) ( -31.50) >DroEre_CAF1 160411 101 - 1 UUUCAUUAC---GGGUCCCCAAA----------AAAGAUUCGCGGGUCGCCAGCCGAAAAGCGACGCCUUGGAAUUCCA--AGGUAUUGGACGUGGAAUUUUGGUUUUCAUCCGUG ......(((---((((..(((((----------(....((((((.(((((..........)))))(((((((....)))--))))......)))))).)))))).....))))))) ( -34.60) >DroYak_CAF1 158347 111 - 1 UCUCAUUACGACGGGUCCCCAAAAAAAA---AAAAAGAUUCGCGGGUUGCCAGCCAAAGAGCGACGCCUUGGAAUUCCA--AGGUGUUGGACGUGGAUUUUUGGUUUUCAUCCGUG ..........((((((........((((---..((((((((((((((.....)))......(((((((((((....)))--))))))))..)))))))))))..)))).)))))). ( -38.90) >DroAna_CAF1 171434 107 - 1 UCUUAUUAC---GGGUUCCCGAAAAUAAAAUAAAAACCUGCGAGGGUUGUCAACCAGAGCGCGACGCCUUGGAAUGCCAAAAGGUGUCGGCCGUGGGUUUUCGGC------CCGUG ......(((---(((...((((((((........(((((....))))).....(((..((.(((((((((((....)))..))))))))))..))))))))))))------))))) ( -43.60) >consensus UCUCAUUAC___GGGUCCCCAAA__________AAAGAUUCGCGGGUCGCCAGCCAAAAUGCGACGCCUUGGAAUUCCA__AGGUGUUGGACGUGGAUUUUUGGUUUUCAGCCGUG ............((....)).............((((((((((((((.....)))......(((((((((((....)))..))))))))..)))))))))))((.......))... (-22.99 = -24.43 + 1.45)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:19:06 2006