| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,518,788 – 14,518,887 |
| Length | 99 |
| Max. P | 0.842911 |

| Location | 14,518,788 – 14,518,887 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 78.54 |
| Mean single sequence MFE | -24.23 |
| Consensus MFE | -9.71 |
| Energy contribution | -11.05 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.40 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.842911 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14518788 99 - 20766785 -CGAAUACAGCAAAAGCAACAGGCAGUAUCGUCAGGCAGCCCAAAAAAGUAGGCAACACUUUU-UGCUGCAUCUCGAUUU-ACUC--AAAAAAAAUUGGCUGAA -......((((....((.....))(((((((....(((((...(((((((.(....)))))))-))))))....)))..)-))).--...........)))).. ( -26.20) >DroSec_CAF1 151111 97 - 1 -CGAAUACAGCAAAAGCAACAGGCAGUAUCGUCAGGCAGCCCAAAAAAGUAGGCAACACUUUU-UGCUGCAGCUUGCUUU-ACUC--AAAAAAA--UGGCUGAA -......((((.((((((((.(((......)))..(((((...(((((((.(....)))))))-)))))).).)))))))-...(--(......--)))))).. ( -27.70) >DroSim_CAF1 152263 97 - 1 -CGAAUACAGCAAAAGCAACAGGCAGUAUCGUCAGGCAGCCCAAAAAAGUAGGCAACACUUUU-UGCUGCAGUUCGCUUU-GCAC--AAAAAAA--UGGCUUAA -........(((((.(((((.(((......)))..(((((...(((((((.(....)))))))-)))))).))).)))))-))..--.......--........ ( -26.10) >DroEre_CAF1 156611 95 - 1 ---AGUACAGCAAAAGCAACAGGCAGUAUCGUCC-GCAGCCCAAAAAAGUAGGCAACACUUUU-UGCUGCAGCUUGUUUU-ACUUU-AAAAAAA--UGCCUGAA ---......((....))..(((((..........-(((((...(((((((.(....)))))))-)))))).....(((((-.....-...))))--)))))).. ( -25.00) >DroYak_CAF1 154967 99 - 1 -CGAAUACAGCAAAAGCAACAGGCAUUAUCAUCG-GAAGCCCAAAAAAGUAGGCAACACUUUUUUCCUACAGCUUGCUUU-ACUUUCAAAAAAA--UGACUCAA -.(((...((((..(((....(((.((.......-)).))).((((((((.(....)))))))))......)))))))..-...))).......--........ ( -16.80) >DroAna_CAF1 165898 87 - 1 UCCAGAAAAGCGAAAGCAUCAGGCGGUGUGG-----C-----AGUGAGGCAGGCAACACUUAU-UGCUGCUCUUUGAUUUUAC----AAAAAAA--GAAGUAUA .........((....))((((((.((.(..(-----(-----((((((...(....).)))))-)))..))))))))).....----.......--........ ( -23.60) >consensus _CGAAUACAGCAAAAGCAACAGGCAGUAUCGUCA_GCAGCCCAAAAAAGUAGGCAACACUUUU_UGCUGCAGCUUGCUUU_ACUC__AAAAAAA__UGGCUGAA ............(((((((...((((((................((((((.(....))))))).))))))...)))))))........................ ( -9.71 = -11.05 + 1.33)


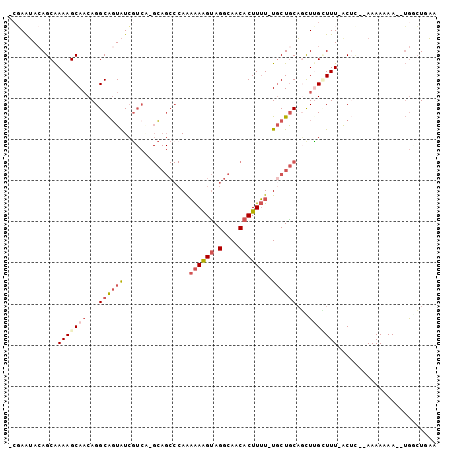
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:19:03 2006