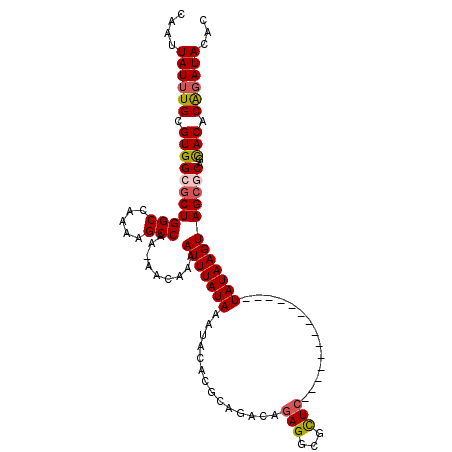
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,509,380 – 14,509,475 |
| Length | 95 |
| Max. P | 0.622549 |

| Location | 14,509,380 – 14,509,475 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 81.57 |
| Mean single sequence MFE | -27.98 |
| Consensus MFE | -20.65 |
| Energy contribution | -20.77 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.583205 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
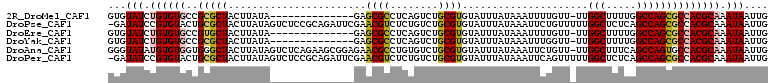
>2R_DroMel_CAF1 14509380 95 + 20766785 GUGUAUCUGUGUGCCGCGCUACUUAUA--------------GAGCGCCUCAGUCUGCGUGUAUUUAUAAAUUUUGUU-UUGGCUUUUGGCCAGCGCCACGCAAAUAAUUG ...(((.((((((..(((((..(((((--------------((((((........))))...)))))))........-..(((.....)))))))))))))).))).... ( -26.70) >DroPse_CAF1 138874 109 + 1 -GAUAUCCGUGUACUGCGCUACUUAUAGUCUCCGCAGAUUCGAACGUCUCUGUCUGCGUGUAUUUAUAAAUUCUGUUUUUGGCUCUCAGCCAGCGCCACGCAAAUAAUUG -.......((((...(((((..((((((...((((((((..((.....)).))))))).)...))))))...........(((.....)))))))).))))......... ( -28.80) >DroEre_CAF1 146935 95 + 1 GUGUAUCUGUGUGCCGUGCUACUUAUA--------------GAGCGCCUCAGUCUGCGUGUAUUUAUAAAUUUUGUU-UUGGCUUUUGGCCAGCGCCACGCAAAUAAUUG ...(((.((((((..(((((..(((((--------------((((((........))))...)))))))........-..(((.....)))))))))))))).))).... ( -24.80) >DroYak_CAF1 145198 95 + 1 GUGUAUCUGUGUGCCGCGCUACUUAUA--------------GAGCGCCUCAGUCUGCGUGUAUUUAUAAAUUUGGUU-UUGGCUUUUGGCCAGCGCCACGCAAAUAAUUG ...(((.((((((.(((((((.(((((--------------((((((........))))...)))))))...)))).-.((((.....))))))).)))))).))).... ( -26.90) >DroAna_CAF1 155450 109 + 1 GGGUAUAUGUGUGGUGGGCUACUUAUAGUCUCAGAAGCGGAGAACGCCUGUGUCUGCGUGUAUUUAUAAAUUCUGUU-UUGGCUUUCAGCCAGUGCCACGCAAAUAAUUG ...(((.((((((((.((((......((((.((((((((((..((....)).)))))(((....)))...)))))..-..))))...))))...)))))))).))).... ( -31.90) >DroPer_CAF1 141076 109 + 1 -GAUAUCCGUGUACUGCGCUACUUAUAGUCUCCGCAGAUUCGAACGUCUCUGUCUGCGUGUAUUUAUAAAUUCAGUUUUUGGCUCUCAGCCAGCGCCACGCAAAUAAUUG -.......((((...(((((..((((((...((((((((..((.....)).))))))).)...))))))...........(((.....)))))))).))))......... ( -28.80) >consensus GGGUAUCUGUGUGCCGCGCUACUUAUA______________GAACGCCUCAGUCUGCGUGUAUUUAUAAAUUCUGUU_UUGGCUUUCAGCCAGCGCCACGCAAAUAAUUG ...(((.((((((..(((((.......................((((........)))).....................(((.....)))))))))))))).))).... (-20.65 = -20.77 + 0.11)
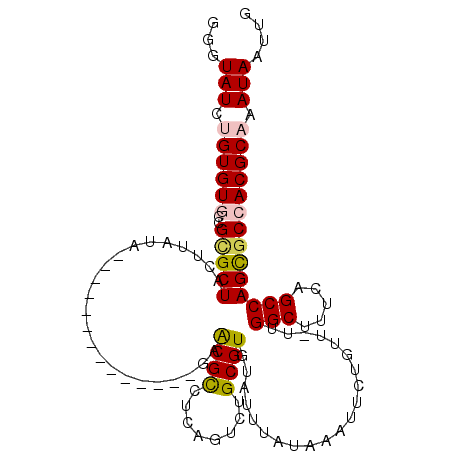

| Location | 14,509,380 – 14,509,475 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 81.57 |
| Mean single sequence MFE | -25.13 |
| Consensus MFE | -16.78 |
| Energy contribution | -16.92 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.622549 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14509380 95 - 20766785 CAAUUAUUUGCGUGGCGCUGGCCAAAAGCCAA-AACAAAAUUUAUAAAUACACGCAGACUGAGGCGCUC--------------UAUAAGUAGCGCGGCACACAGAUACAC ....((((((.(((.(((((((.....)))).-......(((((((......(((........)))...--------------)))))))...))).))).))))))... ( -24.80) >DroPse_CAF1 138874 109 - 1 CAAUUAUUUGCGUGGCGCUGGCUGAGAGCCAAAAACAGAAUUUAUAAAUACACGCAGACAGAGACGUUCGAAUCUGCGGAGACUAUAAGUAGCGCAGUACACGGAUAUC- ....((((((.(((((((((((.....))).........(((((((......((((((..(((...)))...)))))).....))))))))))))..))).))))))..- ( -29.50) >DroEre_CAF1 146935 95 - 1 CAAUUAUUUGCGUGGCGCUGGCCAAAAGCCAA-AACAAAAUUUAUAAAUACACGCAGACUGAGGCGCUC--------------UAUAAGUAGCACGGCACACAGAUACAC ....((((((.(((.((.((((.....)))).-......(((((((......(((........)))...--------------)))))))....)).))).))))))... ( -20.80) >DroYak_CAF1 145198 95 - 1 CAAUUAUUUGCGUGGCGCUGGCCAAAAGCCAA-AACCAAAUUUAUAAAUACACGCAGACUGAGGCGCUC--------------UAUAAGUAGCGCGGCACACAGAUACAC ....((((((.(((.(((((((.....)))).-......(((((((......(((........)))...--------------)))))))...))).))).))))))... ( -24.80) >DroAna_CAF1 155450 109 - 1 CAAUUAUUUGCGUGGCACUGGCUGAAAGCCAA-AACAGAAUUUAUAAAUACACGCAGACACAGGCGUUCUCCGCUUCUGAGACUAUAAGUAGCCCACCACACAUAUACCC ....(((.((.((((...((((.....)))).-......(((((((.....((((........))))((((.......)))).)))))))......)))).)).)))... ( -21.40) >DroPer_CAF1 141076 109 - 1 CAAUUAUUUGCGUGGCGCUGGCUGAGAGCCAAAAACUGAAUUUAUAAAUACACGCAGACAGAGACGUUCGAAUCUGCGGAGACUAUAAGUAGCGCAGUACACGGAUAUC- ....((((((.(((((((((((.....))).........(((((((......((((((..(((...)))...)))))).....))))))))))))..))).))))))..- ( -29.50) >consensus CAAUUAUUUGCGUGGCGCUGGCCAAAAGCCAA_AACAAAAUUUAUAAAUACACGCAGACAGAGGCGCUC______________UAUAAGUAGCGCAGCACACAGAUACAC ....((((((.(((((((((((.....))).........(((((((..............(((...)))..............))))))))))))..))).))))))... (-16.78 = -16.92 + 0.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:19:00 2006