| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,504,807 – 14,504,972 |
| Length | 165 |
| Max. P | 0.538374 |

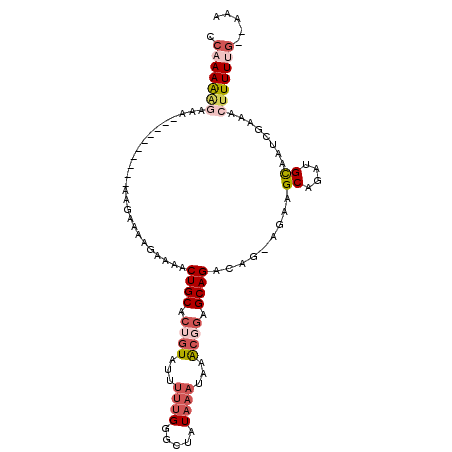
| Location | 14,504,807 – 14,504,910 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 79.08 |
| Mean single sequence MFE | -17.61 |
| Consensus MFE | -9.27 |
| Energy contribution | -10.85 |
| Covariance contribution | 1.58 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.538374 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14504807 103 - 20766785 CCAAAAAGA-A-AAACAAAUAAAGAAAUGAAAACUGCACUGUAUUUUUGGGCUAUAAAUAAACGGAGCAGACGGGAGAAGCAGAUGCAAUCGAAACUUUUUG--AAA .(((((((.-.-.....................((((.((((...((((.....))))...)))).))))....((...((....))..))....)))))))--... ( -17.90) >DroSec_CAF1 136913 104 - 1 CCAAAAAGAAAAAAACAAAUCAAGAAAAGAAAACUGCACUGUAUUUUUGGGCUAUAAAUAAACGGAGCAGGCGG-CGGAGCAGAUGUAAUCGAAACUUUUUG--AUA .(((((((..........(((............((((.((((...((((.....))))...)))).))))((..-....)).)))..........)))))))--... ( -19.15) >DroSim_CAF1 138050 103 - 1 CCAAAAAGGAA-AAACAAAUCAAGAAAAGAAAACUGCACUGUAUUUUUGGGCUAUAAAUAAACGGAGCAGACGG-CGAAGCAGAUGCAAUCGAAACUUUUUG--AAA .(((((((...-.....................((((.((((...((((.....))))...)))).))))....-(((.((....))..)))...)))))))--... ( -18.80) >DroEre_CAF1 142399 92 - 1 CCAAAAGGAA------------AAAAAAGAAAACUGCACGGUAUUUUUGGGCUAUAAAUAAACGGAGCAGACAG-AGGAGCAGAUGCAAUCGAAACUUUUGG--AAA (((((((...------------...........((((.(.((...((((.....))))...)).).))))...(-(...((....))..))....)))))))--... ( -18.30) >DroYak_CAF1 140497 95 - 1 CCAAAAGCAAAG---------AAAAAAAGAAAACUGCACUGUAUUUUUGGGCUAUAAAUAAACGGAGCAGACAG-AGAAGCAGAUGCAAUCGAAACUUUUCG--AAA ............---------.............((((((((.((((((.(((............)))...)))-))).)))).)))).(((((....))))--).. ( -19.60) >DroAna_CAF1 150205 85 - 1 CCAAAG---------------------AUAGAACUGCAGAAUAUUCGAGGGCUAUAAAUAAGCGAAGCAGACAA-AACAGCAGAUGCAAUCGAAACUUUUUCGGAAA ((...(---------------------((....((((.((....))....(((.......)))...))))....-....((....)).)))(((.....)))))... ( -11.90) >consensus CCAAAAAGAAA__________AAGAAAAGAAAACUGCACUGUAUUUUUGGGCUAUAAAUAAACGGAGCAGACAG_AGAAGCAGAUGCAAUCGAAACUUUUUG__AAA .(((((((.........................((((.((((...((((.....))))...)))).)))).........((....))........)))))))..... ( -9.27 = -10.85 + 1.58)

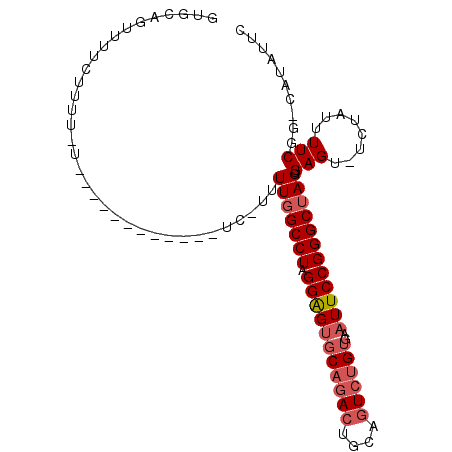
| Location | 14,504,873 – 14,504,972 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 69.86 |
| Mean single sequence MFE | -22.48 |
| Consensus MFE | -14.02 |
| Energy contribution | -15.80 |
| Covariance contribution | 1.78 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.62 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.522523 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14504873 99 + 20766785 GUGCAGUUUUCAUUUCUUUAUUUGUUUU-UCUUUUUGGCCUAGGAGUGCAGACUGCAGUCUGUGAAUUCCGGGCUAGUGAGU-UCUAUUUUCGG-CAUAUUC ((((........................-.....(((((((.(((((((((((....))))))..))))))))))))((((.-......)))))-))).... ( -26.00) >DroEre_CAF1 142464 89 + 1 GUGCAGUUUUCUUUUUU-----------UUCCUUUUGGCCUAGGAGUGCAGACUGCAGUCUGUGAAUUCCGGGCUAGUGAGU-UCUAUUUUCGG-UAUAUUC ((((.............-----------......(((((((.(((((((((((....))))))..))))))))))))((((.-......)))))-))).... ( -24.00) >DroAna_CAF1 150272 78 + 1 CUGCAGUUCUAU--------------------CUUUGGCCUCGGGG--C--AGUGCAGUCAGUGAAUUCCGGGCAAGGGAGUUUCUAUUUUCAAUCCCACUC ............--------------------.....(((.((((.--(--(.((....)).))...)))))))..((((..............)))).... ( -17.44) >consensus GUGCAGUUUUCUUUU_U____________UC_UUUUGGCCUAGGAGUGCAGACUGCAGUCUGUGAAUUCCGGGCUAGUGAGU_UCUAUUUUCGG_CAUAUUC ..................................(((((((.(((((((((((....))))))..)))))))))))).(((........))).......... (-14.02 = -15.80 + 1.78)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:18:58 2006