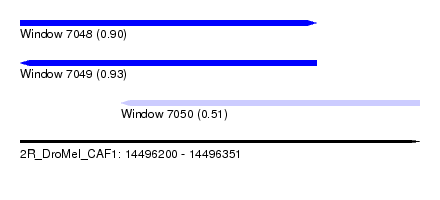
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,496,200 – 14,496,351 |
| Length | 151 |
| Max. P | 0.928032 |

| Location | 14,496,200 – 14,496,312 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 81.26 |
| Mean single sequence MFE | -33.38 |
| Consensus MFE | -21.49 |
| Energy contribution | -22.63 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.904702 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
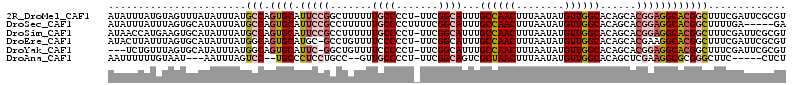
>2R_DroMel_CAF1 14496200 112 + 20766785 AUAUUUAUGUAGUUUAUAUUUAUGCCAGUGCAUUCCGGCUUUUUUGCCCCU-UUCGGCAUUUGCCAACUUUAAUAUGUUGGCACAGCACGGAGGCACGGCUUUCGAUUCGCGU (((..((((.....))))..)))(((.((((.((((((((....((((...-...))))..(((((((........))))))).))).))))))))))))............. ( -37.60) >DroSec_CAF1 128287 108 + 1 AUAUUUAUUUAGUGCAUAUUUAUGCCAGUGCAUUCCGCCUUUUUUGCCCCUUUUCGGCAUUUGCCAACUUUAAUAUGUUGGCACAGCACGGAGGCACGGCUUUUGA-----GA ..................((((.(((.((((.(((((.......((((.......))))..(((((((........))))))).....))))))))))))...)))-----). ( -32.50) >DroSim_CAF1 129419 112 + 1 AUAACCAUGAAGUGCAUAUUUAUGCCAGUGCAUUCCGCCUUUUUUGCCCCU-UUCGGCAUUUGCCAACUUUAAUAUGUUGGCACAGCACGGAGGCACGGCUUUCGAUUCGCGU ...........(((.((......(((.((((.(((((.......((((...-...))))..(((((((........))))))).....)))))))))))).....)).))).. ( -35.30) >DroEre_CAF1 133560 111 + 1 AUACUUAUUUAGUGCAUAUUUAUGGCAGUGCAUGC-GCCUGUUUUCCCCCU-UUCGGCAUUUGCCAACUUUAAUAUGUUGGCACAGCACGAAGGCACGGCUUUCGAUUCGCGU .((((.....))))..........((.((((....-(((............-...)))...(((((((........)))))))..))))((((((...)))))).....)).. ( -29.66) >DroYak_CAF1 131706 108 + 1 ---UCUGUUUAGUGCAUAUUUAUGGCAGUGCAUUC-GGCUGUUUUCCCCCU-UUCGGCAUUUGCCAACUUUAAUAUGUUGGCACAGCACGGAGGCACGGCUUUCGAUUCGCGU ---..........((..(((...(((.((((.(((-((((((......((.-...)).....((((((........))))))))))).)))).)))).)))...)))..)).. ( -34.60) >DroAna_CAF1 141209 100 + 1 AAUUUUUUGUAAU---AAUUUAGUCC--UGCCCUCCUGCC--GUUGCCCCU-UUCGGCAGUCGCUAACUUUAAUAUGUUGGCACAGCUCGAAGGCGCGGGCUUC-----CUCU .............---.....(((((--((((.(((((((--(........-..))))))..((((((........)))))).......)).)))).)))))..-----.... ( -30.60) >consensus AUAUUUAUGUAGUGCAUAUUUAUGCCAGUGCAUUCCGCCUUUUUUGCCCCU_UUCGGCAUUUGCCAACUUUAAUAUGUUGGCACAGCACGGAGGCACGGCUUUCGAUUCGCGU .......................(((.((((.(((((.......((((.......))))...((((((........))))))......))))))))))))............. (-21.49 = -22.63 + 1.14)
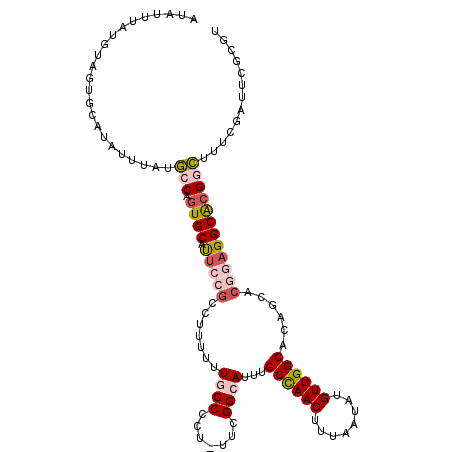

| Location | 14,496,200 – 14,496,312 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 81.26 |
| Mean single sequence MFE | -33.75 |
| Consensus MFE | -19.91 |
| Energy contribution | -20.22 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.20 |
| Mean z-score | -3.11 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.928032 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14496200 112 - 20766785 ACGCGAAUCGAAAGCCGUGCCUCCGUGCUGUGCCAACAUAUUAAAGUUGGCAAAUGCCGAA-AGGGGCAAAAAAGCCGGAAUGCACUGGCAUAAAUAUAAACUACAUAAAUAU .............(((((((.((((.(((.(((((((........)))))))..((((...-...))))....)))))))..)))).)))....................... ( -36.90) >DroSec_CAF1 128287 108 - 1 UC-----UCAAAAGCCGUGCCUCCGUGCUGUGCCAACAUAUUAAAGUUGGCAAAUGCCGAAAAGGGGCAAAAAAGGCGGAAUGCACUGGCAUAAAUAUGCACUAAAUAAAUAU ..-----......(((((((.(((((....(((((((........)))))))..((((.......))))......)))))..)))).)))....................... ( -34.00) >DroSim_CAF1 129419 112 - 1 ACGCGAAUCGAAAGCCGUGCCUCCGUGCUGUGCCAACAUAUUAAAGUUGGCAAAUGCCGAA-AGGGGCAAAAAAGGCGGAAUGCACUGGCAUAAAUAUGCACUUCAUGGUUAU ............((((((((.(((((....(((((((........)))))))..((((...-...))))......)))))..))))..((((....)))).......)))).. ( -36.00) >DroEre_CAF1 133560 111 - 1 ACGCGAAUCGAAAGCCGUGCCUUCGUGCUGUGCCAACAUAUUAAAGUUGGCAAAUGCCGAA-AGGGGGAAAACAGGC-GCAUGCACUGCCAUAAAUAUGCACUAAAUAAGUAU ..(((........((.((((....(((((.(((((((........)))))))..(.((...-.)).).......)))-))..)))).))........)))............. ( -30.99) >DroYak_CAF1 131706 108 - 1 ACGCGAAUCGAAAGCCGUGCCUCCGUGCUGUGCCAACAUAUUAAAGUUGGCAAAUGCCGAA-AGGGGGAAAACAGCC-GAAUGCACUGCCAUAAAUAUGCACUAAACAGA--- ..(((........((.((((...((.(((((((((((........))))))...(.((...-.)).)....))))))-)...)))).))........)))..........--- ( -33.39) >DroAna_CAF1 141209 100 - 1 AGAG-----GAAGCCCGCGCCUUCGAGCUGUGCCAACAUAUUAAAGUUAGCGACUGCCGAA-AGGGGCAAC--GGCAGGAGGGCA--GGACUAAAUU---AUUACAAAAAAUU ....-----..((.((..((((((..((((((((..........((((...)))).((...-.))))).))--)))..)))))).--)).)).....---............. ( -31.20) >consensus ACGCGAAUCGAAAGCCGUGCCUCCGUGCUGUGCCAACAUAUUAAAGUUGGCAAAUGCCGAA_AGGGGCAAAAAAGCCGGAAUGCACUGGCAUAAAUAUGCACUAAAUAAAUAU ................((.(((.((.((..(((((((........)))))))...))))...))).))............((((....))))..................... (-19.91 = -20.22 + 0.31)
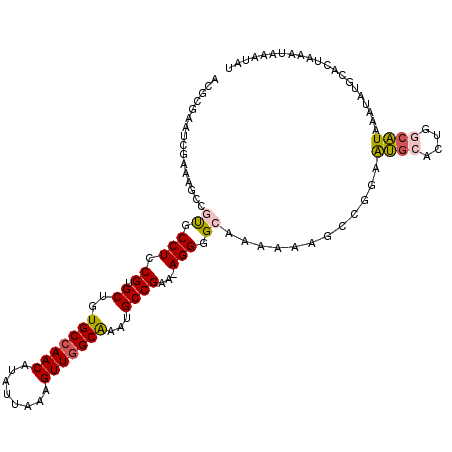
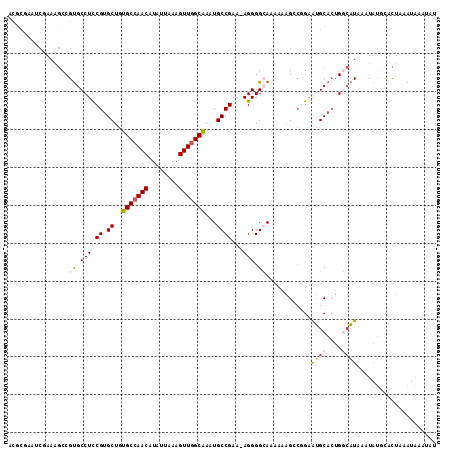
| Location | 14,496,238 – 14,496,351 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 83.87 |
| Mean single sequence MFE | -35.95 |
| Consensus MFE | -22.80 |
| Energy contribution | -23.53 |
| Covariance contribution | 0.73 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.63 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.506987 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
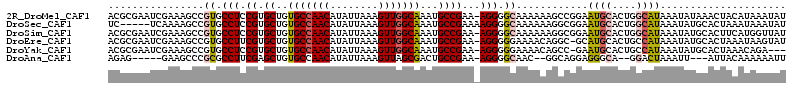
>2R_DroMel_CAF1 14496238 113 - 20766785 UUCAACAUCCGGCUUCCGCUGCAAUGUAAUGGAAACUCGACGCGAAUCGAAAGCCGUGCCUCCGUGCUGUGCCAACAUAUUAAAGUUGGCAAAUGCCGAA-AGGGGCAAAAA-AG .........((((((.((.(((...((...(....)...)))))...)).))))))((((((((.((..(((((((........)))))))...))))..-.))))))....-.. ( -37.30) >DroPse_CAF1 127641 109 - 1 UUCAACAUCCGGCUUCCGUUGCAGUGUAAUGGAAACUCAAGGCGA-----GUGCCGUGCCGCCAUGCUGUGCCAACAUAUUAAAGUUGGCAAAUGCCGAA-AGCACCAGAACCAG (((.......((((((((((((...)))))))))((((.....))-----)))))((((((.(((....(((((((........))))))).))).))..-.))))..))).... ( -35.70) >DroSec_CAF1 128325 109 - 1 UUCAACAUCCGGCUUCCGCUGCAAUGUAAUGGAAACUCGUC-----UCAAAAGCCGUGCCUCCGUGCUGUGCCAACAUAUUAAAGUUGGCAAAUGCCGAAAAGGGGCAAAAA-AG .........((((((....((..(((....(....).))).-----.)).))))))((((((((.((..(((((((........)))))))...))))....))))))....-.. ( -32.90) >DroEre_CAF1 133597 113 - 1 UUCAACAUCCGGCUUCCGUUGCAAUGUAAUGGAAACUCGACGCGAAUCGAAAGCCGUGCCUUCGUGCUGUGCCAACAUAUUAAAGUUGGCAAAUGCCGAA-AGGGGGAAAAC-AG .......(((((((((((((((...)))))))))..((((......))))..)))...((((((.((..(((((((........)))))))...)))).)-))).)))....-.. ( -40.10) >DroAna_CAF1 141242 106 - 1 UUCAACAUCCGGCUUCCGCUACAAUGUAAUGGAAACUCAAGAG-----GAAGCCCGCGCCUUCGAGCUGUGCCAACAUAUUAAAGUUAGCGACUGCCGAA-AGGGGCAAC---GG ........(((((((((((......))...(....)......)-----)))))).((.((((((.((..(((.(((........))).)))...)))).)-))).))...---)) ( -34.00) >DroPer_CAF1 129799 109 - 1 UUCAACAUCCGGCUUCCGUUGCAGUGUAAUGGAAACUCAAGGCGA-----GUGCCGUGCCGCCAUGCUGUGCCAACAUAUUAAAGUUGGCAAAUGCCGAA-AGCACCAGAACCAG (((.......((((((((((((...)))))))))((((.....))-----)))))((((((.(((....(((((((........))))))).))).))..-.))))..))).... ( -35.70) >consensus UUCAACAUCCGGCUUCCGCUGCAAUGUAAUGGAAACUCAACGCGA___GAAAGCCGUGCCUCCGUGCUGUGCCAACAUAUUAAAGUUGGCAAAUGCCGAA_AGGGGCAAAAC_AG .........(((((((((.(((...))).)))))..................))))((((((((.((..(((((((........)))))))...))))....))))))....... (-22.80 = -23.53 + 0.73)

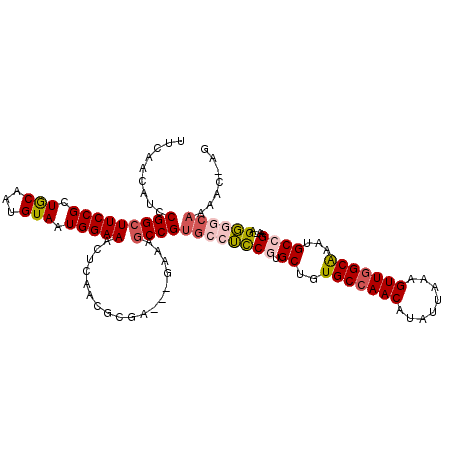
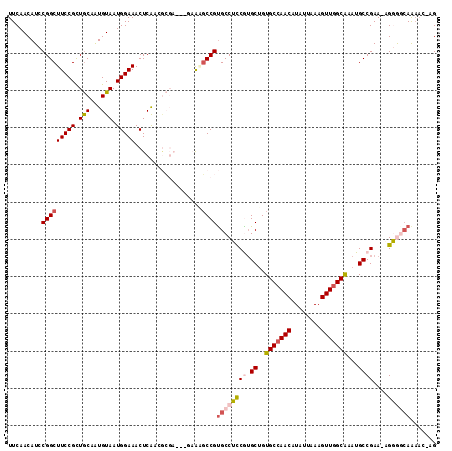
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:18:57 2006