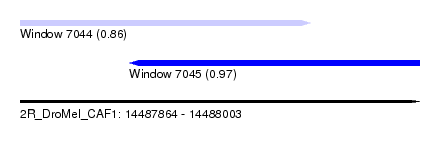
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,487,864 – 14,488,003 |
| Length | 139 |
| Max. P | 0.969761 |

| Location | 14,487,864 – 14,487,965 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 75.61 |
| Mean single sequence MFE | -25.73 |
| Consensus MFE | -13.11 |
| Energy contribution | -14.70 |
| Covariance contribution | 1.59 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.860085 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14487864 101 + 20766785 AAAAUUAACUGAGAUAUAGUGCGAUUUCUGG--CCCGAAAAUUGUGCUAUUUUACAGAACAGGCCACUGACCACCACAUAGACCGCAAGAGAAUACGGAUAAU ........(((.((.((((..(((((((...--...).))))))..)))).)).)))..(((....))).............(((..........)))..... ( -18.60) >DroGri_CAF1 137853 100 + 1 AUAAAUAAUUACAUUAUUGUGAAAUAGAAGCAUACCGUUA---GUGACAAAUCGCUGGCCAUAGAAGUGGCCAGCGAAUGCGCUACAGCAGCAAGCGAGAAAU ........(((((....))))).............((((.---((......(((((((((((....))))))))))).(((......))))).))))...... ( -31.20) >DroSec_CAF1 119710 101 + 1 AAAAUUAACAGAGAUAUAGUGCGAUUUCUGG--CCCGAAAAUUGUGCUAUAUCACGGGACAGGCCACUGACCACCACAUAGACCGCAAGGGAAUACGGAUAAU ............(((((((..(((((((...--...).))))))..))))))).((((.(((....))).))..........((....)).....))...... ( -27.60) >DroSim_CAF1 120864 101 + 1 AAAAUUAACAGAGAUAUAGUGCGAUUUCUGG--CCCGAAAAUUGUGCUAUAUCACGGGACAGGCCACUGACCACCACAUAGACCGCAAGGGAAUACGGAUAAU ............(((((((..(((((((...--...).))))))..))))))).((((.(((....))).))..........((....)).....))...... ( -27.60) >DroEre_CAF1 121377 101 + 1 CAAAAUAGCUAAUAUAUAGUGCGAAUUCUGA--CCCGAAAAUAGUGCUAUAUCACGGGGCAGGCCACUGACCACCAUAUAGACCGCAAGGGAAUACAGAUGAU ........(((..((((((..(...(((...--...)))....)..))))))...(((((((....))).)).))...))).((....))............. ( -22.70) >DroYak_CAF1 123136 101 + 1 CUAAAUAGCAGCGAUAUAGUGCGAAUUCUGG--CCUGAAAAUAGUGCUAUAUCACGGGGCAGACCACUGACCACCAAAUAGACCGCAAGGGAAUACGGAUAAU (((.......(.(((((((..(...(((...--...)))....)..))))))).)(((((((....))).)).))...))).((....))............. ( -26.70) >consensus AAAAAUAACAGAGAUAUAGUGCGAUUUCUGG__CCCGAAAAUUGUGCUAUAUCACGGGACAGGCCACUGACCACCACAUAGACCGCAAGGGAAUACGGAUAAU ............((((((((((..((((.(....).))))...))))))))))...((.(((....))).))..........((....))............. (-13.11 = -14.70 + 1.59)

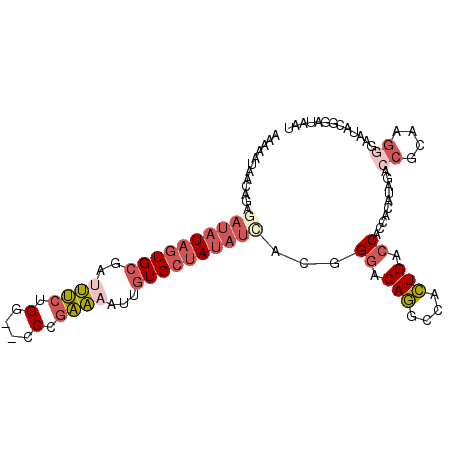
| Location | 14,487,902 – 14,488,003 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 77.18 |
| Mean single sequence MFE | -34.99 |
| Consensus MFE | -19.10 |
| Energy contribution | -19.05 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.46 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.55 |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.969761 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14487902 101 - 20766785 CCUCGUU--UUCCAGCGGCCUUUUCCGGGCUAGGGGACGCAUUAUCCGUAUUCUCUUGCGGUCUAUGUGGUGGUCAGUGGCCUGUUCUGUAAAAUAGCACAAU ((((((.--.....))(((((.....))))).)))).(((((...(((((......)))))...)))))..(((.....)))(((.((((...)))).))).. ( -28.40) >DroGri_CAF1 137893 99 - 1 CGUCUUUUUUUCCAU-GGCAUUUAAUAUGUCAAUGUGCACAUUUCUCGCUUGCUGCUGUAGCGCAUUCGCUGGCCACUUCUAUGGCCAGCGAUUUGUCAC--- ..............(-(((((.....))))))(((((((((......((.....))))).))))))((((((((((......))))))))))........--- ( -32.50) >DroSec_CAF1 119748 101 - 1 CCUCGUU--UUCCAGCGGCCUUUUCCGGGACAGUGGCCGCAUUAUCCGUAUUCCCUUGCGGUCUAUGUGGUGGUCAGUGGCCUGUCCCGUGAUAUAGCACAAU ...(((.--.....)))((..(.(((((((((...(((((((...(((((......)))))...)))))))(((.....)))))))))).)).)..))..... ( -37.90) >DroSim_CAF1 120902 101 - 1 CCUCGUU--UUCCAGCGGCCUUUUCCGGGACAGUGGCCGCAUUAUCCGUAUUCCCUUGCGGUCUAUGUGGUGGUCAGUGGCCUGUCCCGUGAUAUAGCACAAU ...(((.--.....)))((..(.(((((((((...(((((((...(((((......)))))...)))))))(((.....)))))))))).)).)..))..... ( -37.90) >DroEre_CAF1 121415 101 - 1 CGUCGUU--UUGCAACGACCUUUUCCGGGCCAGUGGCCACAUCAUCUGUAUUCCCUUGCGGUCUAUAUGGUGGUCAGUGGCCUGCCCCGUGAUAUAGCACUAU .((((((--....))))))......(((((((.(((((((.....(((((......)))))........))))))).)))))))....(((......)))... ( -38.02) >DroYak_CAF1 123174 101 - 1 CGUCGUU--UUCCAACGGCCUUUUAUGGGCCAGUGGCCACAUUAUCCGUAUUCCCUUGCGGUCUAUUUGGUGGUCAGUGGUCUGCCCCGUGAUAUAGCACUAU .((((((--....)))))).......((((((.(((((((.....(((((......)))))........))))))).)))))).....(((......)))... ( -35.22) >consensus CCUCGUU__UUCCAGCGGCCUUUUCCGGGCCAGUGGCCACAUUAUCCGUAUUCCCUUGCGGUCUAUGUGGUGGUCAGUGGCCUGUCCCGUGAUAUAGCACAAU ..(((((......))))).......((((.((.(((((((.....(((((......)))))........))))))).)).))))....(((......)))... (-19.10 = -19.05 + -0.05)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:18:52 2006