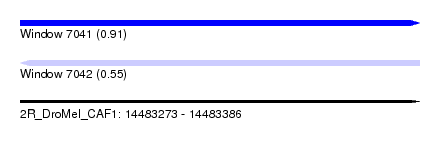
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,483,273 – 14,483,386 |
| Length | 113 |
| Max. P | 0.912716 |

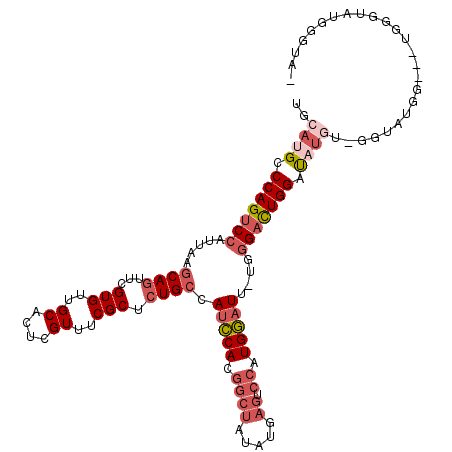
| Location | 14,483,273 – 14,483,386 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 81.39 |
| Mean single sequence MFE | -32.79 |
| Consensus MFE | -22.99 |
| Energy contribution | -24.27 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.912716 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14483273 113 + 20766785 AUACCCAUACCCA---CCAUACCCACAUAUCCAGUCCCA-AAUCCAUGGACUCAUAUAGCCGUGGAUGGCAGAGCGAAACGAGUGCAACACGAACUGCUUAAUGGACUGGGCAUGCA .............---.........(((..(((((((..-.((((((((.((.....))))))))))(((((..((..............))..)))))....)))))))..))).. ( -33.94) >DroSec_CAF1 115114 112 + 1 -UACCCAUAGCCA---CCAUACCCACAUAUCCAGUCCCA-AAUCCAUGGACUCAUAUAGCCGUGGAUGGCAGAGCGAAACAAGUGCAACACGAACUGCUUAAUGGACUGGGCAUGCA -............---.........(((..(((((((..-.((((((((.((.....))))))))))(((((..((..............))..)))))....)))))))..))).. ( -33.94) >DroSim_CAF1 116258 112 + 1 -UACCCAUAGCCA---CCAUACCUACAUAUCCAGUCCCA-AAUCCAGGGACUCAUAUAGCCGUGGAUGGCAGAGCGAAACGAGUGCAACACGAACUGCUUAAUGGACUGGGCAUGCA -............---.........(((..(((((((..-.(((((.((.((.....)))).)))))(((((..((..............))..)))))....)))))))..))).. ( -32.14) >DroEre_CAF1 116796 106 + 1 --ACCCAUGCCCAAGCCAAU--------AUCCAGUCCCA-AAUCCAUGGACUCACAUAGCCGUGGAUGGCAGAGCGAAACGAGUGCAACACGAACUGCUUAAUGGACUGGGCAUGCA --...((((((((..(((.(--------(..((((.(..-.((((((((.((.....)))))))))).(((...((...))..))).....).))))..)).)))..)))))))).. ( -37.10) >DroYak_CAF1 118426 113 + 1 --ACCCAUACCCAAGCCAAUACC--CAUAUCCAGUCCAAAAAUCCAUGGACUCAUAUAGCCGUGGAUGGCAGAGCGAAACGAGUGCAACACGAACUGCUUAAUGGACUGGGCAUGCA --.....................--(((..((((((((...((((((((.((.....))))))))))(((((..((..............))..)))))...))))))))..))).. ( -35.64) >DroAna_CAF1 128205 101 + 1 -AACCUU-----------AAACCAAAAAGCCCAAACGCA-UAUACAUGC---CUGGCACCUAUGCCUGGCAUCGCGAAACGAGUGCAACACGAACUGUUUAAUGGACUGGCCAUGGA -......-----------...(((...((.(((...(((-(.....(((---(.((((....)))).))))(((.....)))))))((((.....))))...))).)).....))). ( -24.00) >consensus _UACCCAUACCCA___CCAUACC_ACAUAUCCAGUCCCA_AAUCCAUGGACUCAUAUAGCCGUGGAUGGCAGAGCGAAACGAGUGCAACACGAACUGCUUAAUGGACUGGGCAUGCA ..............................(((((((....((((((((.((.....))))))))))(((((..((..............))..)))))....)))))))....... (-22.99 = -24.27 + 1.28)

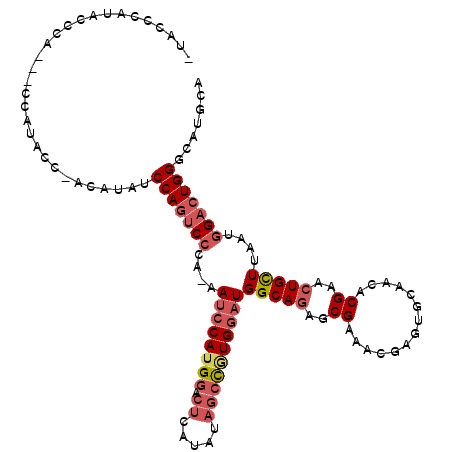

| Location | 14,483,273 – 14,483,386 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 81.39 |
| Mean single sequence MFE | -37.27 |
| Consensus MFE | -23.92 |
| Energy contribution | -25.70 |
| Covariance contribution | 1.78 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.554489 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14483273 113 - 20766785 UGCAUGCCCAGUCCAUUAAGCAGUUCGUGUUGCACUCGUUUCGCUCUGCCAUCCACGGCUAUAUGAGUCCAUGGAUU-UGGGACUGGAUAUGUGGGUAUGG---UGGGUAUGGGUAU ...(((((((..((((((.((((...(((..((....))..))).)))).(((((((..(((.(.(((((.......-..))))).))))))))))).)))---)))...))))))) ( -38.00) >DroSec_CAF1 115114 112 - 1 UGCAUGCCCAGUCCAUUAAGCAGUUCGUGUUGCACUUGUUUCGCUCUGCCAUCCACGGCUAUAUGAGUCCAUGGAUU-UGGGACUGGAUAUGUGGGUAUGG---UGGCUAUGGGUA- ....((((((..((((((.((((...(((..((....))..))).)))).(((((((..(((.(.(((((.......-..))))).))))))))))).)))---)))...))))))- ( -37.20) >DroSim_CAF1 116258 112 - 1 UGCAUGCCCAGUCCAUUAAGCAGUUCGUGUUGCACUCGUUUCGCUCUGCCAUCCACGGCUAUAUGAGUCCCUGGAUU-UGGGACUGGAUAUGUAGGUAUGG---UGGCUAUGGGUA- ....((((((..(((....((((...(((..((....))..))).))))...(((.(.(((((((((((((......-.))))))...))))))).).)))---)))...))))))- ( -38.30) >DroEre_CAF1 116796 106 - 1 UGCAUGCCCAGUCCAUUAAGCAGUUCGUGUUGCACUCGUUUCGCUCUGCCAUCCACGGCUAUGUGAGUCCAUGGAUU-UGGGACUGGAU--------AUUGGCUUGGGCAUGGGU-- ..(((((((((.(((.((..(((((((((..((....))..)))......(((((.((((.....)).)).))))).-..))))))..)--------).))).)))))))))...-- ( -39.60) >DroYak_CAF1 118426 113 - 1 UGCAUGCCCAGUCCAUUAAGCAGUUCGUGUUGCACUCGUUUCGCUCUGCCAUCCACGGCUAUAUGAGUCCAUGGAUUUUUGGACUGGAUAUG--GGUAUUGGCUUGGGUAUGGGU-- ..(((((((((.(((....((((......))))((((((.((.....(((......)))......((((((........)))))).)).)))--)))..))).)))))))))...-- ( -39.80) >DroAna_CAF1 128205 101 - 1 UCCAUGGCCAGUCCAUUAAACAGUUCGUGUUGCACUCGUUUCGCGAUGCCAGGCAUAGGUGCCAG---GCAUGUAUA-UGCGUUUGGGCUUUUUGGUUU-----------AAGGUU- .((..((((((((((...(((...(((((..((....))..)))))((((.((((....)))).)---)))......-...))))))))....))))).-----------..))..- ( -30.70) >consensus UGCAUGCCCAGUCCAUUAAGCAGUUCGUGUUGCACUCGUUUCGCUCUGCCAUCCACGGCUAUAUGAGUCCAUGGAUU_UGGGACUGGAUAUGU_GGUAUGG___UGGGUAUGGGUA_ ..((((.(((((((.....((((...(((..((....))..))).)))).(((((.((((.....)).)).)))))....))))))).))))......................... (-23.92 = -25.70 + 1.78)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:18:49 2006