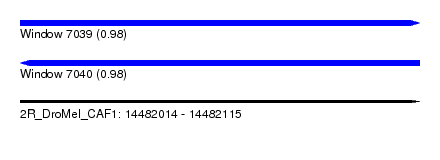
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,482,014 – 14,482,115 |
| Length | 101 |
| Max. P | 0.982795 |

| Location | 14,482,014 – 14,482,115 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 78.18 |
| Mean single sequence MFE | -25.05 |
| Consensus MFE | -13.60 |
| Energy contribution | -15.18 |
| Covariance contribution | 1.58 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.92 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.92 |
| SVM RNA-class probability | 0.982795 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14482014 101 + 20766785 -GCUGACGCUUCAUCGCUGCCAGCC-AAGAAA-UGCAAUUUUCGGCAGCAGCAGCAGCAACAGCAA-CUGCAAGGCAAAAAUGCAUAAAACGGAUGAAAA-AAUAU -((....))((((((((((((....-..((((-......)))))))))).((((..((....))..-))))...(((....)))........))))))..-..... ( -27.80) >DroSec_CAF1 113923 99 + 1 -GCUGACGCUUCAUCGCUGCCAGCC-AAGAAA-UGCAAUUUUCGGCAGCAGCAG---CAGAAGCAA-CUGCAAGGCAAAAAUGCAUAAAACGGAUGAAAAAAAAAU -((....))(((((((((((..(((-..((((-......))))))).))))).(---(((......-))))...(((....)))........))))))........ ( -28.20) >DroSim_CAF1 115020 101 + 1 -GCUGACGCUUCAUCGCUGCCAGCC-AAGAAA-UGCAAUUUUCGGCAGCAGCAGCAGCAAAAGCAA-CUGCAAGGCAAAAAUGCAUAAAACGGAUGAAAA-AAAAU -((....))((((((((((((....-..((((-......)))))))))).((((..((....))..-))))...(((....)))........))))))..-..... ( -27.80) >DroEre_CAF1 115614 96 + 1 UGCUGAUGCCUCAUCGCUGCCAGCC-AAGAAA-UGCAAUUUUCGGCAGCAGCAACAGCG---GCUACAUGGC----AAAAAUGCAUAGGAGGGAGGGGAA-AAAAU ..((..(.((((...((((((((((-..((((-......)))))))(((.((....)).---)))...))))----).....))....)))).)..))..-..... ( -26.10) >DroYak_CAF1 117165 98 + 1 UGAUGAUGCUUCAUCGCUGCCAGCC-AAGAAAUUGCAAUUUUCGGCAGCAGCAACAGCA---ACAA-AUGCAAAA-AAAAAUGCAUAAGACAGAGG-AAA-AAAAU .(((((....)))))(((((..(((-..((((.......))))))).))))).......---....-(((((...-.....)))))..........-...-..... ( -23.30) >DroPer_CAF1 114154 87 + 1 -GCUGAAGCUUUAUCGCUUCCAGCAAAAGCAA-UGCAAUUUUUGGCAGCAGCAAAAACAAAAA-----CAC-AGAAAAAAAUGC----------UGGAAA-AAAAU -((((((((......)))).))))........-.....(((((..(((((.............-----...-.........)))----------))..))-))).. ( -17.11) >consensus _GCUGACGCUUCAUCGCUGCCAGCC_AAGAAA_UGCAAUUUUCGGCAGCAGCAACAGCAAAAGCAA_CUGCAAGGCAAAAAUGCAUAAAACGGAUGAAAA_AAAAU .........(((((((((((..(((.((((........)))).))).)))))................((((.........)))).......))))))........ (-13.60 = -15.18 + 1.58)

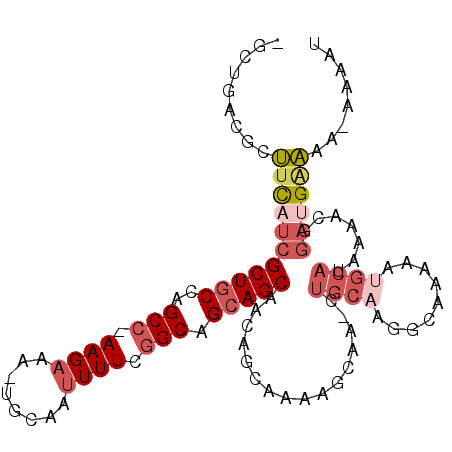
| Location | 14,482,014 – 14,482,115 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 78.18 |
| Mean single sequence MFE | -27.98 |
| Consensus MFE | -14.14 |
| Energy contribution | -14.78 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.51 |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.975670 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
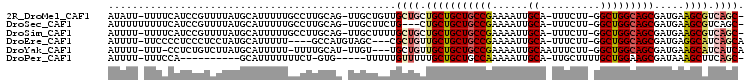
>2R_DroMel_CAF1 14482014 101 - 20766785 AUAUU-UUUUCAUCCGUUUUAUGCAUUUUUGCCUUGCAG-UUGCUGUUGCUGCUGCUGCUGCCGAAAAUUGCA-UUUCUU-GGCUGGCAGCGAUGAAGCGUCAGC- .....-........(((((((((((....)))...((((-(.......)))))((((((((((((.((.....-.)).))-))).))))))))))))))).....- ( -31.10) >DroSec_CAF1 113923 99 - 1 AUUUUUUUUUCAUCCGUUUUAUGCAUUUUUGCCUUGCAG-UUGCUUCUG---CUGCUGCUGCCGAAAAUUGCA-UUUCUU-GGCUGGCAGCGAUGAAGCGUCAGC- ..............(((((((((((....)))...((((-......)))---)((((((((((((.((.....-.)).))-))).))))))))))))))).....- ( -29.80) >DroSim_CAF1 115020 101 - 1 AUUUU-UUUUCAUCCGUUUUAUGCAUUUUUGCCUUGCAG-UUGCUUUUGCUGCUGCUGCUGCCGAAAAUUGCA-UUUCUU-GGCUGGCAGCGAUGAAGCGUCAGC- .....-........(((((((((((....)))...((((-(.......)))))((((((((((((.((.....-.)).))-))).))))))))))))))).....- ( -31.10) >DroEre_CAF1 115614 96 - 1 AUUUU-UUCCCCUCCCUCCUAUGCAUUUUU----GCCAUGUAGC---CGCUGUUGCUGCUGCCGAAAAUUGCA-UUUCUU-GGCUGGCAGCGAUGAGGCAUCAGCA .....-................(((....)----))......((---(..(((((((((((((((.((.....-.)).))-))).)))))))))).)))....... ( -27.80) >DroYak_CAF1 117165 98 - 1 AUUUU-UUU-CCUCUGUCUUAUGCAUUUUU-UUUUGCAU-UUGU---UGCUGUUGCUGCUGCCGAAAAUUGCAAUUUCUU-GGCUGGCAGCGAUGAAGCAUCAUCA .....-...-..........(((((.....-...)))))-.((.---((((((((((((((((((((((....)))).))-))).)))))))))..)))).))... ( -28.10) >DroPer_CAF1 114154 87 - 1 AUUUU-UUUCCA----------GCAUUUUUUUCU-GUG-----UUUUUGUUUUUGCUGCUGCCAAAAAUUGCA-UUGCUUUUGCUGGAAGCGAUAAAGCUUCAGC- .....-....((----------(((.........-(((-----(....((((((((....).))))))).)))-)......)))))(((((......)))))...- ( -19.96) >consensus AUUUU_UUUUCAUCCGUUUUAUGCAUUUUUGCCUUGCAG_UUGCUUUUGCUGCUGCUGCUGCCGAAAAUUGCA_UUUCUU_GGCUGGCAGCGAUGAAGCGUCAGC_ ................................................((((.(((((((((((......((..........))))))))).....)))).)))). (-14.14 = -14.78 + 0.64)
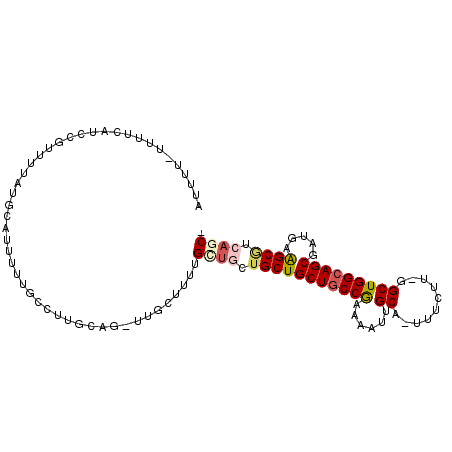
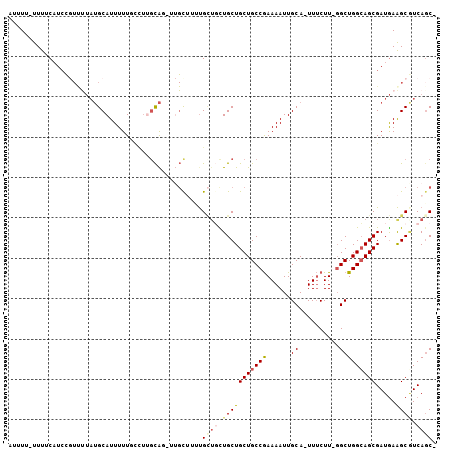
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:18:47 2006