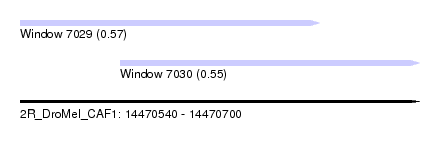
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,470,540 – 14,470,700 |
| Length | 160 |
| Max. P | 0.574140 |

| Location | 14,470,540 – 14,470,660 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.61 |
| Mean single sequence MFE | -43.05 |
| Consensus MFE | -33.00 |
| Energy contribution | -32.83 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.574140 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
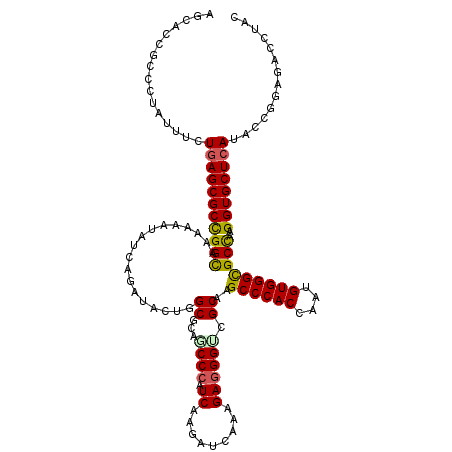
>2R_DroMel_CAF1 14470540 120 + 20766785 AGCACCGCCCUAUUUCUGAGCGCCGGCAAGAAGUACCAGAUCCUGGCCCAGCCCAUCAAGAUCAAGGAGGGUCGCAAGCCCACCAAUGUGGGCGCCAAGGUGCUCAUCCCGGAGACCUAC ................(((((((((((.....(..((((...))))..).((((((.........((.(((.(....)))).))...)))))))))..))))))))....(....).... ( -41.00) >DroVir_CAF1 128940 120 + 1 AGCACGGCUCUCUUUUUGAGCGCUGGCAAAAAAUAUCAGAUACUGGCGCAGCCCAUCAAGAUCAAGGAGGGCCGCAAGCCCACUAAUGUGGGCGCCAAGGUGCUCAUACCGGAGACAUAC .((.((((.(((.....))).))))))................((((((.((((..(........)..)))).))..((((((....)))))))))).((((....))))(....).... ( -47.90) >DroGri_CAF1 120627 120 + 1 AGCACGGCGCUAUUUCUGAGCGCUGGCAAAAAGUAUCAGAUACUGGCUCAGCCCAUCAAGAUCAAAGAGGGACGCAAGCCCACAAAUGUGGGCGCCAAGGUGCUCAUACCGGAGACCUAC .((.(((((((.......)))))))))....(((((...)))))(((...((((.((.........))))).)....((((((....))))))))).((((.(((......))))))).. ( -44.10) >DroWil_CAF1 170088 120 + 1 AGUACGGCAUUAUUUCUAAGCGCUGGUAAAAAAUAUCAAAUCCUAGCGCAACCCAUCAAGAUCAAAGAGGGUCGCAAGCCCACCAAUGUGGGUGCCAAGGUGCUCAUACCCGAGACCUAU .....(((...........(((((((................))))))).((((.((.........)))))).....((((((....))))))))).((((.(((......))))))).. ( -36.59) >DroMoj_CAF1 163503 120 + 1 AGCACCGCGCUGUUCUUGAGCGCCGGCAAGAAAUAUCAGAUACUGGCGCAGCCCAUCAAGAUCAAAGAGGGACGCAAGCCCACCAAUGUGGGCGCUAAGGUGCUCAUACCGGAGACCUAU (((((((((((.(..(((((((((((................)))))))....(.....).))))..).)).)))..((((((....)))))).....))))))......(....).... ( -43.09) >DroAna_CAF1 115003 120 + 1 AGCACCGCCCUGUUCCUGAGCGCCGGCAAGAAGUACCAGAUCCUGGCCCAGCCGAUCAAGAUCAAGGAGGGUCGCAAGCCCACCAACGUGGGCGCCAAGGUGCUCAUCCCAGAAACCUAC .........(((....(((((((((((.....(((((...(((((((...)))(((....))).)))).))).))..((((((....)))))))))..))))))))...)))........ ( -45.60) >consensus AGCACCGCCCUAUUUCUGAGCGCCGGCAAAAAAUAUCAGAUACUGGCGCAGCCCAUCAAGAUCAAAGAGGGUCGCAAGCCCACCAAUGUGGGCGCCAAGGUGCUCAUACCGGAGACCUAC ................(((((((((((..................((...((((.((.........)))))).))..((((((....)))))))))..)))))))).............. (-33.00 = -32.83 + -0.16)

| Location | 14,470,580 – 14,470,700 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.67 |
| Mean single sequence MFE | -46.00 |
| Consensus MFE | -37.66 |
| Energy contribution | -37.00 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.34 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.554277 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
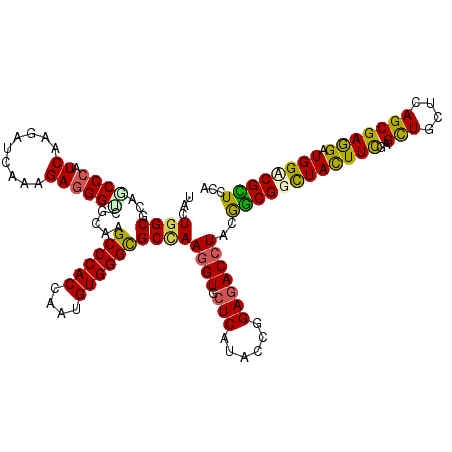
>2R_DroMel_CAF1 14470580 120 + 20766785 UCCUGGCCCAGCCCAUCAAGAUCAAGGAGGGUCGCAAGCCCACCAAUGUGGGCGCCAAGGUGCUCAUCCCGGAGACCUACGGCGGCUACUUCGAGCUCCUCAGCGAGGAUGGUCGCUCCA ....(((...)))............((((((((((..((((((....))))))....((((.(((......)))))))...))))))....(((.((((((...))))).).))))))). ( -45.70) >DroVir_CAF1 128980 120 + 1 UACUGGCGCAGCCCAUCAAGAUCAAGGAGGGCCGCAAGCCCACUAAUGUGGGCGCCAAGGUGCUCAUACCGGAGACAUACGGCGGCUAUUUUGAGCUGCUCAGCGAGGAUGGACGCUCGA ...((((((.((((..(........)..)))).))..((((((....)))))))))).((((..(((.(((....)....(((((((......)))))))......)))))..))))... ( -51.80) >DroGri_CAF1 120667 120 + 1 UACUGGCUCAGCCCAUCAAGAUCAAAGAGGGACGCAAGCCCACAAAUGUGGGCGCCAAGGUGCUCAUACCGGAGACCUACGGCGGCUACUUUGAGCUGCUCAGCGAAGAUGGACGCUCCA ....((...((((((((....((((((.(((.(....)))).......(((.((((.((((.(((......)))))))..)))).)))))))))(((....)))...)))))..))))). ( -45.10) >DroWil_CAF1 170128 120 + 1 UCCUAGCGCAACCCAUCAAGAUCAAAGAGGGUCGCAAGCCCACCAAUGUGGGUGCCAAGGUGCUCAUACCCGAGACCUAUGGCGGUUAUUUUGAGUUACUCAGCGAGGAUGGUCGUUCGA ((((.((((.((((.((.........)))))).))..((((((....))))))((((((((.(((......))))))).))))........((((...)))))).))))........... ( -40.50) >DroMoj_CAF1 163543 120 + 1 UACUGGCGCAGCCCAUCAAGAUCAAAGAGGGACGCAAGCCCACCAAUGUGGGCGCUAAGGUGCUCAUACCGGAGACCUAUAACGGCUACUUCGAGCUGCUUAGCGAGGAUGGCCGUUCGA ...((((((..(((.((.........)))))..))..((((((....))))))))))((((.(((......)))))))..((((((((.(((..(((....)))..)))))))))))... ( -46.20) >DroAna_CAF1 115043 120 + 1 UCCUGGCCCAGCCGAUCAAGAUCAAGGAGGGUCGCAAGCCCACCAACGUGGGCGCCAAGGUGCUCAUCCCAGAAACCUACGGCGGCUACUUCGAGCUACUCAGCGAAGAUGGCCGCUCCA ..((((.......(((....)))..((.(((.(....)))).))...((((((((....)))))))).))))........((((((((((((..(((....))))))).))))))))... ( -46.70) >consensus UACUGGCGCAGCCCAUCAAGAUCAAAGAGGGUCGCAAGCCCACCAAUGUGGGCGCCAAGGUGCUCAUACCGGAGACCUACGGCGGCUACUUCGAGCUGCUCAGCGAGGAUGGACGCUCCA ...((((...((((.((.........)))))).....((((((....))))))))))((((.(((......)))))))..((((((((((((..(((....))))))).))))))))... (-37.66 = -37.00 + -0.66)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:18:37 2006