| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,455,314 – 14,455,423 |
| Length | 109 |
| Max. P | 0.538611 |

| Location | 14,455,314 – 14,455,423 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.59 |
| Mean single sequence MFE | -18.94 |
| Consensus MFE | -14.12 |
| Energy contribution | -13.98 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.538611 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
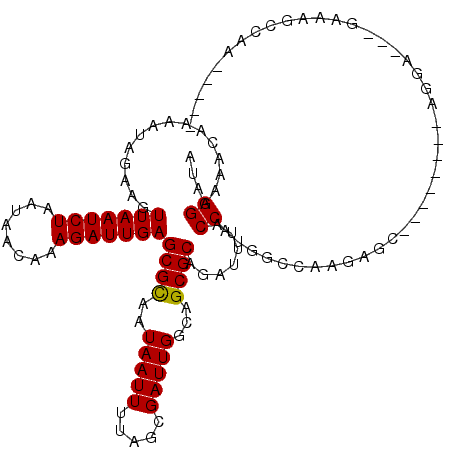
>2R_DroMel_CAF1 14455314 109 + 20766785 AUAGGAAAACAAAAUAGAAGUUAAUCUAAUAACAAAGAUUGAGCGCAAUAAUUUUAGCGAUUGGCAGCGCAGAUUUACCAUGGCCAAGAGCU-------AGGA---GUAAGCCAAGAAG- ...((...((..........(((((((........))))))).........(((((((..(((((.................)))))..)))-------))))---))...))......- ( -21.33) >DroVir_CAF1 112464 96 + 1 AUAGGAAAACAAAAUAGAAGUUAAUCUAAUAACAAAGAUUGAGCGCAAUAAUUUUAGCGAUUGGCAGCGCAGAUUUACCAUGCCGAAGC----------AGGA---GAA----------- ....................(((((((........)))))))((((..(((((.....)))))...)))).......((.(((....))----------))).---...----------- ( -16.30) >DroGri_CAF1 105814 93 + 1 AUAGGAAAACAAAAUAGAAGUUAAUCUAAUAACAAAGAUUGAGCGCAAUAAUUUCAGCGAUUGGCAGCGCAGAUUUACCAUGCUGAA-------------GGA---GAA----------- ...((...............(((((((........)))))))((((..(((((.....)))))...)))).......))........-------------...---...----------- ( -14.40) >DroWil_CAF1 148442 104 + 1 AUAGGAAAACAAAAUAGAAGUUAAUCUAAUAACAAAGAUUGAGCGUAAUAAUUUUAGCGAUUGGCAGCGCCGAUUUACCAUGGGCAAAAAUC-------G--C---AAAAAAUAUA---- ....................(((((((........))))))).....(((.((((.((((((......(((.((.....)).)))...))))-------)--)---.)))).))).---- ( -17.00) >DroYak_CAF1 90080 117 + 1 AUAGGAAAACAAAAUAGAAGUUAAUCUAAUAACAAAGAUUGAGCGCAAUAAUUUUAGCGAUUGGCAGCGCAGAUUUACCAUGGCCAAGAGCUAGGAGCUAGGA---GUAAGCCAAGCAAA ....................(((((((........))))))).........(((((((..(((((.................)))))..)))))))(((.((.---.....)).)))... ( -23.73) >DroAna_CAF1 98751 109 + 1 AUAGGAAAACAAAAUAGAAGUUAAUCUAAUAACAAAGAUUGAGCGCAAUAAUUUCAGCGAUUGGCAGCGCAGAUUUACCAUGGCCAAGAGCC-------AACUUCCAACAGCCAAC---- ...((((.............(((((((........)))))))((((..(((((.....)))))...))))..........((((.....)))-------)..))))..........---- ( -20.90) >consensus AUAGGAAAACAAAAUAGAAGUUAAUCUAAUAACAAAGAUUGAGCGCAAUAAUUUUAGCGAUUGGCAGCGCAGAUUUACCAUGGCCAAGAGC________AGGA___GAAAGCCAA_____ ...((...............(((((((........)))))))((((..(((((.....)))))...)))).......))......................................... (-14.12 = -13.98 + -0.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:18:34 2006