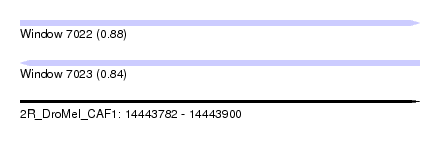
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,443,782 – 14,443,900 |
| Length | 118 |
| Max. P | 0.883141 |

| Location | 14,443,782 – 14,443,900 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 81.05 |
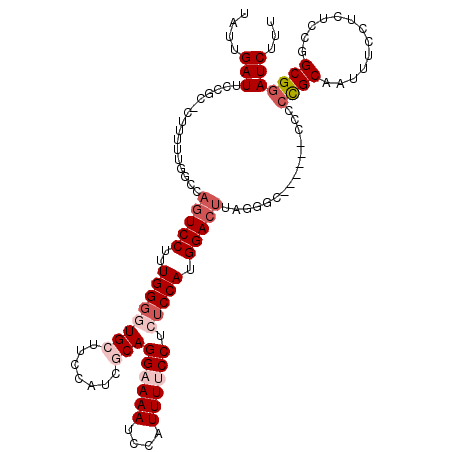
| Mean single sequence MFE | -34.89 |
| Consensus MFE | -20.32 |
| Energy contribution | -21.32 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.883141 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14443782 118 + 20766785 AAAGAUCCGCCGGAGAGGAAAUUGCGGGGGGGGGGGGUCCAAAGUCCAUGGAGAGGAAAAUGGAUUUUCCUGCGAUGGAAGCACCCCAAAGGACUGGCCAAAAAGAGCGGAAUCAAUA ...((((((((.....((..(.(.(.(((((((..((((((...(((.......)))...))))))..)))((.......)).))))...).).)..)).....).)))).))).... ( -40.30) >DroSec_CAF1 75812 111 + 1 AAAGAUCCGCCGGAGAGGAAAUUGCGGGGG------GCCCUAAGUCCAUGGAGAGGAAAAUGGAUUUUCCUGCGAUGGAAGCAACCCAAAGGACUGGCCAAAAAG-GCGGAAUCAAUA ...((((((((.....((...(((((((..------..)))...(((((.(..(((((((....))))))).).))))).)))).(((......))))).....)-)))).))).... ( -37.40) >DroSim_CAF1 77098 112 + 1 AAAGAUCCGCCGGAGAGGAAAUUGCAGGGG-----CGCCCUAAGUCCAUGGAGAGGAAAAUGGAUUUUCCUGCGAUGGAAGCAACCCAAAGGACUGGCCAAAAAG-GCGGAAUCAAUA ...((((((((.....((...((((((((.-----..))))...(((((.(..(((((((....))))))).).))))).)))).(((......))))).....)-)))).))).... ( -37.70) >DroEre_CAF1 77348 92 + 1 AAAGAUCCGCUGGGUAGGAAAACGCG--GG------GCUCUAAGUCCAUGGAGAGGAAAACGGAUUUGCCUGUGAUGGUAGCACCCCAAAGGA------------------AUCGGUG ...(((((..((((........((((--((------....(((((((..............))))))))))))).((....)).))))..)).------------------))).... ( -22.04) >DroYak_CAF1 78100 110 + 1 AAAGAUCCGCCGGAGAGGAAAUUGCGGGGG------GUUCCAAGUCCAUGGGGUGGAAAAUGGAUUUGCCUGUGAUGGGAGCACGCCAAAGGACUAGCCAAA-GG-GCGGCAUCAAUG ...((((((((((.......((..((((.(------(.((((..(((((...)))))...)))).)).))))..)).........))...((.....))...-.)-)))).))).... ( -36.99) >consensus AAAGAUCCGCCGGAGAGGAAAUUGCGGGGG______GCCCUAAGUCCAUGGAGAGGAAAAUGGAUUUUCCUGCGAUGGAAGCACCCCAAAGGACUGGCCAAAAAG_GCGGAAUCAAUA ...(((((((..........((((((((((........))(((((((((..........)))))))))))))))))((.......))...................)))).))).... (-20.32 = -21.32 + 1.00)

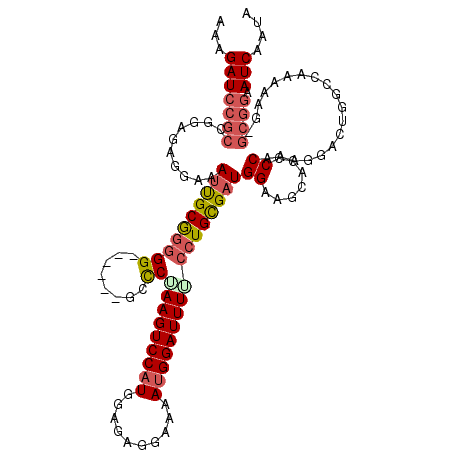
| Location | 14,443,782 – 14,443,900 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 81.05 |
| Mean single sequence MFE | -29.34 |
| Consensus MFE | -15.22 |
| Energy contribution | -17.06 |
| Covariance contribution | 1.84 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.835202 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14443782 118 - 20766785 UAUUGAUUCCGCUCUUUUUGGCCAGUCCUUUGGGGUGCUUCCAUCGCAGGAAAAUCCAUUUUCCUCUCCAUGGACUUUGGACCCCCCCCCCCCGCAAUUUCCUCUCCGGCGGAUCUUU ....(((.(((((......((..(((((..((((((((.......)))((((((....)))))).))))).)))))..((......))............)).....))))))))... ( -29.80) >DroSec_CAF1 75812 111 - 1 UAUUGAUUCCGC-CUUUUUGGCCAGUCCUUUGGGUUGCUUCCAUCGCAGGAAAAUCCAUUUUCCUCUCCAUGGACUUAGGGC------CCCCCGCAAUUUCCUCUCCGGCGGAUCUUU ....(((.((((-(.....((((((....))))(((((.(((((.(.(((((((....)))))))..).)))))....(((.------..))))))))..)).....))))))))... ( -33.80) >DroSim_CAF1 77098 112 - 1 UAUUGAUUCCGC-CUUUUUGGCCAGUCCUUUGGGUUGCUUCCAUCGCAGGAAAAUCCAUUUUCCUCUCCAUGGACUUAGGGCG-----CCCCUGCAAUUUCCUCUCCGGCGGAUCUUU ....(((.((((-(......((((((((..((((.(((.......)))((((((....))))))..)))).)))))...)))(-----(....))............))))))))... ( -34.40) >DroEre_CAF1 77348 92 - 1 CACCGAU------------------UCCUUUGGGGUGCUACCAUCACAGGCAAAUCCGUUUUCCUCUCCAUGGACUUAGAGC------CC--CGCGUUUUCCUACCCAGCGGAUCUUU ....(((------------------((....(((((.(((((((...(((.(((.....))))))....))))...))).))------))--)((.............)))))))... ( -21.52) >DroYak_CAF1 78100 110 - 1 CAUUGAUGCCGC-CC-UUUGGCUAGUCCUUUGGCGUGCUCCCAUCACAGGCAAAUCCAUUUUCCACCCCAUGGACUUGGAAC------CCCCCGCAAUUUCCUCUCCGGCGGAUCUUU ....(((.((((-(.-....(((((....))))).(((..((......))....((((...((((.....))))..))))..------.....)))...........))))))))... ( -27.20) >consensus UAUUGAUUCCGC_CUUUUUGGCCAGUCCUUUGGGGUGCUUCCAUCGCAGGAAAAUCCAUUUUCCUCUCCAUGGACUUAGGGC______CCCCCGCAAUUUCCUCUCCGGCGGAUCUUU ....(((................(((((..((((((((.......)))((((((....)))))).))))).)))))...............((((.............)))))))... (-15.22 = -17.06 + 1.84)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:18:30 2006