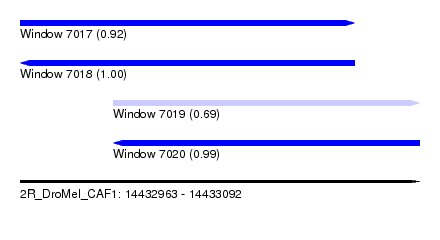
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,432,963 – 14,433,092 |
| Length | 129 |
| Max. P | 0.995811 |

| Location | 14,432,963 – 14,433,071 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.55 |
| Mean single sequence MFE | -21.50 |
| Consensus MFE | -10.10 |
| Energy contribution | -9.60 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.47 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.916776 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
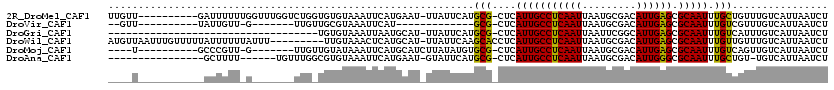
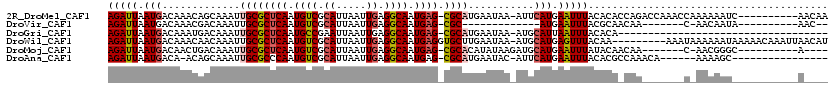
>2R_DroMel_CAF1 14432963 108 + 20766785 UUGUU----------GAUUUUUUGGUUUGGUCUGGUGUGUAAAUUCAUGAAU-UUAUUCAUGCG-CUCAUUGCCUCAAUUAAUGCGACAUUGAGCGCAAUUUGCUGUUUGUCAUUAAUCU ....(----------(((....((((........(((.(((((((....)))-)))).)))(((-((((.((.(.((.....)).).)).))))))).....))))...))))....... ( -22.90) >DroVir_CAF1 85177 86 + 1 --GUU----------UAUUGUU-G-------UUGUUGCGUAAAUUCAU-------------GCG-CUCAUUGCCUCAAUUAAUGCGACAUUGAGCGCAAUUUGUCGUUUGUCAUUAAUCU --...----------....(((-(-------.((..((((((((...(-------------(((-((((.((.(.((.....)).).)).))))))))))))).)))....)).)))).. ( -19.70) >DroGri_CAF1 82309 83 + 1 -----------------------------------UGUGUAAAUUAAUGCAU-UUAUUCAUGCG-CUCAUUGCCUCAAUUAAUUCGGCAUUGAGCGCAAUUUGUCAUUUGUCAUUAAUCU -----------------------------------.......(((((((((.-.....((((((-((((.((((...........)))).))))))))...)).....)).))))))).. ( -24.80) >DroWil_CAF1 115771 110 + 1 AUGUUAAUUUGUUUUUAUUUUUUAUUU---------UUGUAAACUCAUGCAU-UUAUUCAAGCACCUCAUUGCCUCAAUUAAUGCGACAUUGAGCGCAAUUUGUUGUUUGUCAUUAAUCU ..((((((...................---------.((((......)))).-.....((((((.(..(((((((((((.........)))))).)))))..).))))))..)))))).. ( -17.40) >DroMoj_CAF1 106840 97 + 1 ----U----------GCCCGUU-G-------UUGUUGUAUAAAUUCAUGCAUCUUAUAUGUGCG-CUCAUUGCCUCAAUUAAUGCGACAUUGAGCGCAAUUUGUCAGUUGUCAUUAAUCU ----.----------.......-.-------...........(((.(((((.((.(((..((((-((((.((.(.((.....)).).)).))))))))...))).)).)).))).))).. ( -20.20) >DroAna_CAF1 75818 95 + 1 ----------------GCUUUU------UGUUUGGCGUGUAAAUUCAUGAAU-GUAUUCAUGCG-CUCAUUGCCUCAAUUAAUGCGACAUUGGGCGCAAUUUGCUGU-UGUCAUUAAUCU ----------------......------....(((((.(((((((.(((((.-...)))))(((-((((.((.(.((.....)).).)).))))))))))))))...-)))))....... ( -24.00) >consensus ____U___________AUUUUU_G_______UUG_UGUGUAAAUUCAUGCAU_UUAUUCAUGCG_CUCAUUGCCUCAAUUAAUGCGACAUUGAGCGCAAUUUGUCGUUUGUCAUUAAUCU .............................................................(((....(((((((((((.........)))))).))))).)))................ (-10.10 = -9.60 + -0.50)
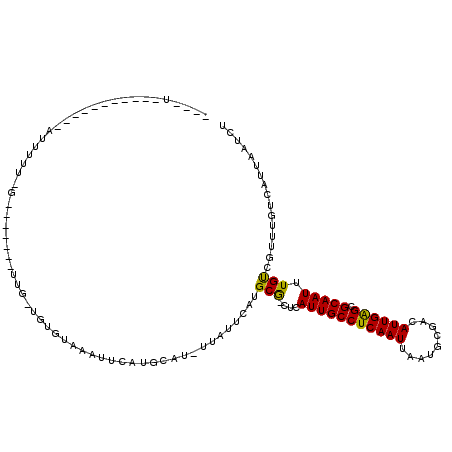
| Location | 14,432,963 – 14,433,071 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.55 |
| Mean single sequence MFE | -21.70 |
| Consensus MFE | -14.81 |
| Energy contribution | -15.23 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.14 |
| Mean z-score | -3.18 |
| Structure conservation index | 0.68 |
| SVM decision value | 2.62 |
| SVM RNA-class probability | 0.995811 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14432963 108 - 20766785 AGAUUAAUGACAAACAGCAAAUUGCGCUCAAUGUCGCAUUAAUUGAGGCAAUGAG-CGCAUGAAUAA-AUUCAUGAAUUUACACACCAGACCAAACCAAAAAAUC----------AACAA .((((.................((((((((.((((.((.....)).)))).))))-))))((..(((-(((....))))))..))................))))----------..... ( -23.70) >DroVir_CAF1 85177 86 - 1 AGAUUAAUGACAAACGACAAAUUGCGCUCAAUGUCGCAUUAAUUGAGGCAAUGAG-CGC-------------AUGAAUUUACGCAACAA-------C-AACAAUA----------AAC-- ......................((((((((.((((.((.....)).)))).))))-)))-------------)................-------.-.......----------...-- ( -20.30) >DroGri_CAF1 82309 83 - 1 AGAUUAAUGACAAAUGACAAAUUGCGCUCAAUGCCGAAUUAAUUGAGGCAAUGAG-CGCAUGAAUAA-AUGCAUUAAUUUACACA----------------------------------- (((((((((.((..........((((((((.((((...........)))).))))-)))).......-.))))))))))).....----------------------------------- ( -26.97) >DroWil_CAF1 115771 110 - 1 AGAUUAAUGACAAACAACAAAUUGCGCUCAAUGUCGCAUUAAUUGAGGCAAUGAGGUGCUUGAAUAA-AUGCAUGAGUUUACAA---------AAAUAAAAAAUAAAAACAAAUUAACAU ...........((((..(..(((((.((((((.........)))))))))))..)((((........-..))))..))))....---------........................... ( -16.20) >DroMoj_CAF1 106840 97 - 1 AGAUUAAUGACAACUGACAAAUUGCGCUCAAUGUCGCAUUAAUUGAGGCAAUGAG-CGCACAUAUAAGAUGCAUGAAUUUAUACAACAA-------C-AACGGGC----------A---- (((((.(((.((.((.......((((((((.((((.((.....)).)))).))))-))))......)).))))).))))).........-------.-.......----------.---- ( -24.72) >DroAna_CAF1 75818 95 - 1 AGAUUAAUGACA-ACAGCAAAUUGCGCCCAAUGUCGCAUUAAUUGAGGCAAUGAG-CGCAUGAAUAC-AUUCAUGAAUUUACACGCCAAACA------AAAAGC---------------- ............-...((.....((((.((.((((.((.....)).)))).)).)-)))(((((...-.)))))..........))......------......---------------- ( -18.30) >consensus AGAUUAAUGACAAACAACAAAUUGCGCUCAAUGUCGCAUUAAUUGAGGCAAUGAG_CGCAUGAAUAA_AUGCAUGAAUUUACACA_CAA_______C_AAAAAU___________A____ (((((.(((.............((((((((.((((.((.....)).)))).)))).))))...........))).)))))........................................ (-14.81 = -15.23 + 0.42)
| Location | 14,432,993 – 14,433,092 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.70 |
| Mean single sequence MFE | -20.19 |
| Consensus MFE | -10.10 |
| Energy contribution | -9.60 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.693334 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14432993 99 + 20766785 AAAUUCAUGAAU-UUAUUCAUGCG-CUCAUUGCCUCAAUUAAUGCGACAUUGAGCGCAAUUUGCUGUUUGUCAUUAAUCUUCUUUU--CA--------------GUUUUUGGUUA---CG ......(((((.-...)))))(((-((((.((.(.((.....)).).)).)))))))((((.((((....................--))--------------))....)))).---.. ( -17.45) >DroVir_CAF1 85197 85 + 1 AAAUUCAU-------------GCG-CUCAUUGCCUCAAUUAAUGCGACAUUGAGCGCAAUUUGUCGUUUGUCAUUAAUCUUCUUUU--CA--------------UUU--UCAUUU---GG .......(-------------(((-((((.((.(.((.....)).).)).))))))))............................--..--------------...--......---.. ( -15.80) >DroGri_CAF1 82314 99 + 1 AAAUUAAUGCAU-UUAUUCAUGCG-CUCAUUGCCUCAAUUAAUUCGGCAUUGAGCGCAAUUUGUCAUUUGUCAUUAAUCUUCUUUUUUCA--------------UUU--UCACUU---GG ..(((((((((.-.....((((((-((((.((((...........)))).))))))))...)).....)).)))))))............--------------...--......---.. ( -24.80) >DroWil_CAF1 115802 103 + 1 AAACUCAUGCAU-UUAUUCAAGCACCUCAUUGCCUCAAUUAAUGCGACAUUGAGCGCAAUUUGUUGUUUGUCAUUAAUCUUCUUAA--GC--------------AUUUUUCUUUGUCUCU ......((((..-.....((((((.(..(((((((((((.........)))))).)))))..).))))))...((((.....))))--))--------------)).............. ( -18.10) >DroMoj_CAF1 106858 100 + 1 AAAUUCAUGCAUCUUAUAUGUGCG-CUCAUUGCCUCAAUUAAUGCGACAUUGAGCGCAAUUUGUCAGUUGUCAUUAAUCUUCUUUG--CA--------------AUUUGUGAUUU---GG ..(((.(((((.((.(((..((((-((((.((.(.((.....)).).)).))))))))...))).)).)).))).))).....(..--(.--------------....)..)...---.. ( -20.60) >DroAna_CAF1 75836 110 + 1 AAAUUCAUGAAU-GUAUUCAUGCG-CUCAUUGCCUCAAUUAAUGCGACAUUGGGCGCAAUUUGCUGU-UGUCAUUAAUCUUCCUUU--CAAGCCGCAAGACUUGGUUUUUGGUUU----- ..(((.((((..-(((....((((-((((.((.(.((.....)).).)).))))))))...)))...-..)))).)))...((..(--((((.(....).))))).....))...----- ( -24.40) >consensus AAAUUCAUGCAU_UUAUUCAUGCG_CUCAUUGCCUCAAUUAAUGCGACAUUGAGCGCAAUUUGUCGUUUGUCAUUAAUCUUCUUUU__CA______________AUUUUUCAUUU___GG .....................(((....(((((((((((.........)))))).))))).)))........................................................ (-10.10 = -9.60 + -0.50)

| Location | 14,432,993 – 14,433,092 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.70 |
| Mean single sequence MFE | -22.82 |
| Consensus MFE | -14.41 |
| Energy contribution | -14.83 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.90 |
| Structure conservation index | 0.63 |
| SVM decision value | 2.32 |
| SVM RNA-class probability | 0.992376 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
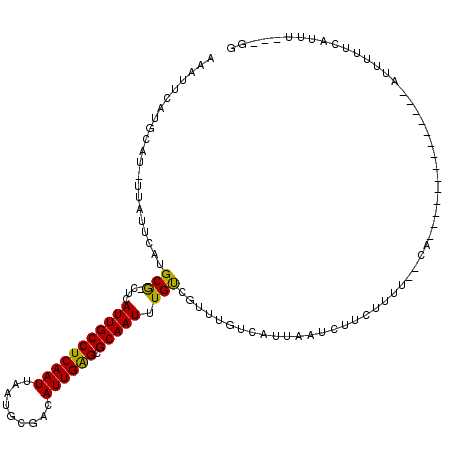
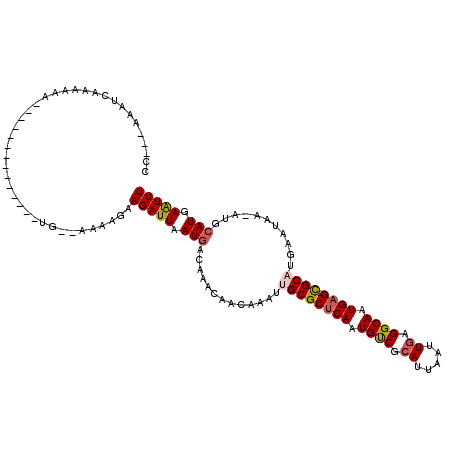
>2R_DroMel_CAF1 14432993 99 - 20766785 CG---UAACCAAAAAC--------------UG--AAAAGAAGAUUAAUGACAAACAGCAAAUUGCGCUCAAUGUCGCAUUAAUUGAGGCAAUGAG-CGCAUGAAUAA-AUUCAUGAAUUU ..---..........(--------------(.--...)).(((((.((((.......((...((((((((.((((.((.....)).)))).))))-)))))).....-..)))).))))) ( -24.04) >DroVir_CAF1 85197 85 - 1 CC---AAAUGA--AAA--------------UG--AAAAGAAGAUUAAUGACAAACGACAAAUUGCGCUCAAUGUCGCAUUAAUUGAGGCAAUGAG-CGC-------------AUGAAUUU ..---......--...--------------..--............................((((((((.((((.((.....)).)))).))))-)))-------------)....... ( -20.30) >DroGri_CAF1 82314 99 - 1 CC---AAGUGA--AAA--------------UGAAAAAAGAAGAUUAAUGACAAAUGACAAAUUGCGCUCAAUGCCGAAUUAAUUGAGGCAAUGAG-CGCAUGAAUAA-AUGCAUUAAUUU ..---......--...--------------..........(((((((((.((..........((((((((.((((...........)))).))))-)))).......-.))))))))))) ( -26.57) >DroWil_CAF1 115802 103 - 1 AGAGACAAAGAAAAAU--------------GC--UUAAGAAGAUUAAUGACAAACAACAAAUUGCGCUCAAUGUCGCAUUAAUUGAGGCAAUGAGGUGCUUGAAUAA-AUGCAUGAGUUU ................--------------((--(((...........((((...................))))(((((.(((.(((((......))))).))).)-)))).))))).. ( -17.51) >DroMoj_CAF1 106858 100 - 1 CC---AAAUCACAAAU--------------UG--CAAAGAAGAUUAAUGACAACUGACAAAUUGCGCUCAAUGUCGCAUUAAUUGAGGCAAUGAG-CGCACAUAUAAGAUGCAUGAAUUU ..---...........--------------..--......(((((.(((.((.((.......((((((((.((((.((.....)).)))).))))-))))......)).))))).))))) ( -24.32) >DroAna_CAF1 75836 110 - 1 -----AAACCAAAAACCAAGUCUUGCGGCUUG--AAAGGAAGAUUAAUGACA-ACAGCAAAUUGCGCCCAAUGUCGCAUUAAUUGAGGCAAUGAG-CGCAUGAAUAC-AUUCAUGAAUUU -----...........((((((....))))))--......(((((.((((..-.........(((((.((.((((.((.....)).)))).)).)-)))).......-..)))).))))) ( -24.15) >consensus CC___AAAUCAAAAAA______________UG__AAAAGAAGAUUAAUGACAAACAACAAAUUGCGCUCAAUGUCGCAUUAAUUGAGGCAAUGAG_CGCAUGAAUAA_AUGCAUGAAUUU ........................................(((((.(((.............((((((((.((((.((.....)).)))).)))).))))...........))).))))) (-14.41 = -14.83 + 0.42)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:18:27 2006