
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,432,343 – 14,432,441 |
| Length | 98 |
| Max. P | 0.507133 |

| Location | 14,432,343 – 14,432,441 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 85.74 |
| Mean single sequence MFE | -34.12 |
| Consensus MFE | -25.60 |
| Energy contribution | -26.02 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.75 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.507133 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14432343 98 - 20766785 CUUUUGGCCGCCAUCAAGUGUGGCCAA-------AAAUCGGCUGGCGCUUU--AAAACCAUCCAUCUGAGCAAUUG-------GGCAGCAGCUAAAUGCUGGCGGUUCGCUGCU .(((((((((((.....).))))))))-------))...(((.((((....--..((((.....((..(....)..-------))(((((......)))))..))))))))))) ( -33.40) >DroSec_CAF1 64341 98 - 1 UUUUUGGCCGCCAUCAAGUGUGGCCAA-------AAAUCGGCUGGCGCUUU--AAAACCAUCCAUCUGAGCAAUCG-------GGCAGCAGCUAAAUGCUGGCGGUUCGCUGCU ((((((((((((.....).))))))))-------)))..(((.((((....--..((((.....(((((....)))-------))(((((......)))))..))))))))))) ( -36.40) >DroSim_CAF1 65281 98 - 1 CUUUUGGCCGCCAUCAAGUGUGGCCAA-------AAAUCGGCUGGCGCUUU--AAAACCAUCCAUCUGAGCAAUCG-------GGCAGCAGCUAAAUGCUGGCGGUUCGCUGCU .(((((((((((.....).))))))))-------))...(((.((((....--..((((.....(((((....)))-------))(((((......)))))..))))))))))) ( -35.80) >DroEre_CAF1 66131 98 - 1 CUUUUGGCCGCCAUCAAGUGUGGCCAA-------AAAUCGGUUGGCGUUUU--AAAACCGUCCAUCUGAGCAAUCG-------GGCAGCAGCUAAAUGCUGGCGGUUCGCUGUU .(((((((((((.....).))))))))-------)).......((((....--..((((((...(((((....)))-------))(((((......)))))))))))))))... ( -36.20) >DroYak_CAF1 66542 100 - 1 CUUUUGGCCGCCAUCAAGUGUGGCCAA-------AAAUCGGUUGGCGUUUUCAAAAACCAUCCAUCUGAACAAUCG-------GGCAGCAGCUAAAUGCUGGCGGUUCGCAGCU .(((((((((((.....).))))))))-------)).((((.(((.((((....))))...))).)))).....((-------(((.(((((.....))).)).)))))..... ( -32.00) >DroAna_CAF1 75108 105 - 1 CG-UUGGCCACCAUUAAGUGUGGCCAAAAAAAAAAAAUCGGUUAACGGCUG--AAAACCAUCCAUCCGAACAAUCAGCCAACAGGCAGCGGCUAAAUGCUGGCGGUGC------ ..-(((((((((.....).)))))))).........(((.......(((((--(...................))))))....(.(((((......))))).))))..------ ( -30.91) >consensus CUUUUGGCCGCCAUCAAGUGUGGCCAA_______AAAUCGGCUGGCGCUUU__AAAACCAUCCAUCUGAGCAAUCG_______GGCAGCAGCUAAAUGCUGGCGGUUCGCUGCU ...(((((((((.....).))))))))................((((........((((........((....))........(.(((((......))))).)))))))))... (-25.60 = -26.02 + 0.42)
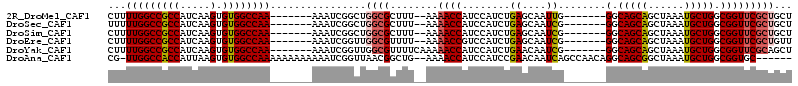

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:18:24 2006