
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,420,097 – 14,420,192 |
| Length | 95 |
| Max. P | 0.594819 |

| Location | 14,420,097 – 14,420,192 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 72.67 |
| Mean single sequence MFE | -28.08 |
| Consensus MFE | -10.47 |
| Energy contribution | -10.38 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.37 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.594819 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14420097 95 + 20766785 -AAGCUGACAAAUUUAUUGUCGCCGGUGACAUAAAUAUCGGUAUCAGAGCU--CAGAGCUC--AGACU-UCAGAAUGCUGACCC--ACU-GGCGAUGUGUCG--CG-- -..((.((((........((((((((((..................((((.--....))))--.....-((((....))))..)--)))-)))))).)))))--).-- ( -30.80) >DroVir_CAF1 71746 89 + 1 -AAGCUGACAAAUUUAUUGUCGCCGACGACAUAAA--UCGGUAUCGGAGCU--GAC---C----------UACAAUUCGCUGCCAGUCU-GGAAAUGUGUCAGAGACA -...((((((.((((..(((((....)))))....--..((((.((((..(--(..---.----------..)).)))).)))).....-..)))).))))))..... ( -22.70) >DroSec_CAF1 52179 95 + 1 -AAGCUGACAAAUUUAUUGUCGCCGGUGACAUAAAUAUCGGUAUCAGAGCU--CAGAGCUC--AGACA-UCAGAAUGCUGACCC--ACU-GGCGAUGUGUCG--CG-- -..((.((((........((((((((((..................((((.--....))))--.....-((((....))))..)--)))-)))))).)))))--).-- ( -30.90) >DroEre_CAF1 53865 96 + 1 -AAGCUGACAAAUUUAUUGUCGCCGGUGAUAUAAAUAUCGGUAUCAGAGCU--CAGAGCUC--AGACU-UCAGAAUUCUGACCC--ACUCGGCGAUGUGUCG--CG-- -..((.((((........((((((((((..................((((.--....))))--.....-((((....))))..)--)).))))))).)))))--).-- ( -30.00) >DroWil_CAF1 100740 106 + 1 AAAGCUGACAAAUUUAUUGUCGCCGUUGACAUAAAUAUCGGUAACAGUCGUAACCGAGUAGAGAGACAGGCAGAGAAUUGACCCA-ACU-GGAAAUGUGUCAGACAGA ...(((((((.(((((((.((((((((....((....(((((.((....)).))))).))....))).))).)).)))....((.-...-)))))).)))))).)... ( -26.50) >DroAna_CAF1 61804 85 + 1 -AAGCUGACAAAUUUAUUGUCGCCGGUGACAUAAAUAUCGGUAUCACUGCU--UCUGGC-----------------AGUGGCCC--ACU-GGAAAUGUGUCAGACGAC -..(((((((.((((((.((((....)))))))))).(((((.(((((((.--....))-----------------)))))...--)))-)).....)))))).)... ( -27.60) >consensus _AAGCUGACAAAUUUAUUGUCGCCGGUGACAUAAAUAUCGGUAUCAGAGCU__CAGAGCUC__AGAC__UCAGAAUACUGACCC__ACU_GGAAAUGUGUCA__CG__ .....(((((.(((..(((..(((((((.......)))))))..)))...................................((......)).))).)))))...... (-10.47 = -10.38 + -0.08)

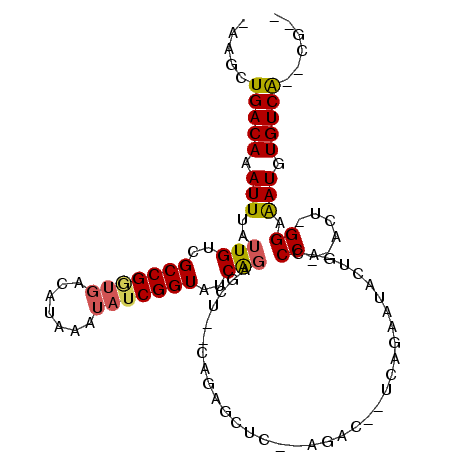
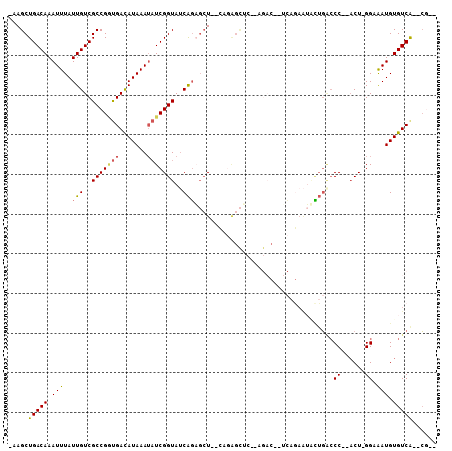
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:18:21 2006