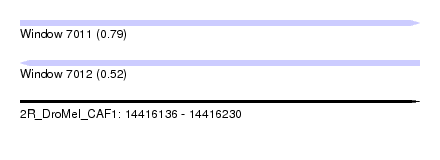
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,416,136 – 14,416,230 |
| Length | 94 |
| Max. P | 0.791022 |

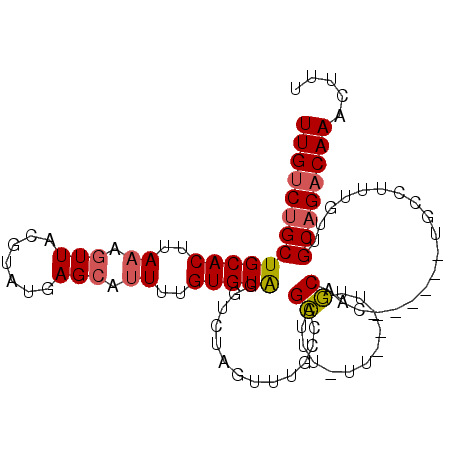
| Location | 14,416,136 – 14,416,230 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 83.93 |
| Mean single sequence MFE | -20.15 |
| Consensus MFE | -13.89 |
| Energy contribution | -15.12 |
| Covariance contribution | 1.22 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.04 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.791022 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14416136 94 + 20766785 AAAGUUUGUCUGCAACAAAGGCA------GUUGCAACAAA--AAAGCUGGCAAACUAGACCUACACAAAAUGCUCAUACGUAACUUUAAGUGCAGCAGACAA ...(((((.(((((..((((.((------(((........--..))))).....................(((......))).))))...)))))))))).. ( -19.70) >DroSec_CAF1 48306 94 + 1 AAAGUUUGUC-GCAACAAAGGCA------GUUGCAACAAAA-AAAGCUGGCAAACUAGACCUACACAAAAUGCUCAUACGUAACUUUAAGUGCAGCAGACAA .....(((((-(((((.......------))))).......-...((((.((...((((..(((...............)))..))))..)))))).))))) ( -18.36) >DroSim_CAF1 51064 96 + 1 AAAGUUUGUCUGCAACAAAGGCA------GUUGCAACAAAAAAAAGCUGGCAAACUAGACCUACACAAAAUGCUCAUACGUAACUUUAAGUGCAGCAGACAA ...(((((.(((((..((((...------((.(((...........((((....))))............)))....))....))))...)))))))))).. ( -20.50) >DroEre_CAF1 50098 91 + 1 AAAGUUUGUCUGCAACAAAGGCA------GUUGCAA---AAA--AGCCAGCAAACUAGACCUACACAAAAUGCUCAUACGUAACUUUAAGUGCAGCAGACAA .....((((((((......(((.------.((....---)).--.))).(((...((((..(((...............)))..))))..))).)))))))) ( -20.46) >DroWil_CAF1 95620 83 + 1 AAAGUUUG-CUGCACU-----CA------CCUGAAA---GU---ACC-CUAAAAAAAUACCCACACAAAGCUCUCAUAUGUAACUUUAAGUGCAGCAGACAA ...(((((-(((((((-----((------..(((.(---((---...-.....................))).)))..))........)))))))))))).. ( -20.66) >DroYak_CAF1 49883 99 + 1 AAAGUUUGUCUGCAACAAAGGCAAAAAAUGUUGCAA---AAAACAGCUAGCAAACUAGACCUACACAAAAUGCUCAUACGUAACUUUAAGUGCAGCAGACAA ...(((((.(((((..((((((((......))))..---.......((((....)))).........................))))...)))))))))).. ( -21.20) >consensus AAAGUUUGUCUGCAACAAAGGCA______GUUGCAA___AA_AAAGCUGGCAAACUAGACCUACACAAAAUGCUCAUACGUAACUUUAAGUGCAGCAGACAA ...(((((.(((((..(((((((........)))............((((....)))).........................))))...)))))))))).. (-13.89 = -15.12 + 1.22)

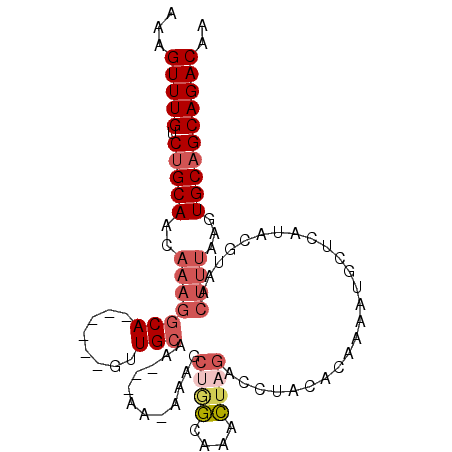
| Location | 14,416,136 – 14,416,230 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 83.93 |
| Mean single sequence MFE | -24.58 |
| Consensus MFE | -15.16 |
| Energy contribution | -15.42 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.62 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.516095 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14416136 94 - 20766785 UUGUCUGCUGCACUUAAAGUUACGUAUGAGCAUUUUGUGUAGGUCUAGUUUGCCAGCUUU--UUUGUUGCAAC------UGCCUUUGUUGCAGACAAACUUU (((((((((((((..((.(((.......))).))..)))))(((..((((.((.(((...--...))))))))------))))......))))))))..... ( -25.90) >DroSec_CAF1 48306 94 - 1 UUGUCUGCUGCACUUAAAGUUACGUAUGAGCAUUUUGUGUAGGUCUAGUUUGCCAGCUUU-UUUUGUUGCAAC------UGCCUUUGUUGC-GACAAACUUU ......(((((((..((.(((.......))).))..)))).(((.......))))))...-.(((((((((((------.......)))))-)))))).... ( -22.60) >DroSim_CAF1 51064 96 - 1 UUGUCUGCUGCACUUAAAGUUACGUAUGAGCAUUUUGUGUAGGUCUAGUUUGCCAGCUUUUUUUUGUUGCAAC------UGCCUUUGUUGCAGACAAACUUU (((((((((((((..((.(((.......))).))..)))))(((..((((.((.(((........))))))))------))))......))))))))..... ( -25.70) >DroEre_CAF1 50098 91 - 1 UUGUCUGCUGCACUUAAAGUUACGUAUGAGCAUUUUGUGUAGGUCUAGUUUGCUGGCU--UUU---UUGCAAC------UGCCUUUGUUGCAGACAAACUUU (((((((((((((..((.(((.......))).))..)))))(((..((((.((.....--...---..)))))------))))......))))))))..... ( -23.40) >DroWil_CAF1 95620 83 - 1 UUGUCUGCUGCACUUAAAGUUACAUAUGAGAGCUUUGUGUGGGUAUUUUUUUAG-GGU---AC---UUUCAGG------UG-----AGUGCAG-CAAACUUU ..((.((((((((((((((((.(......))))))).((..(((((((.....)-)))---))---)..))..------.)-----)))))))-)).))... ( -25.90) >DroYak_CAF1 49883 99 - 1 UUGUCUGCUGCACUUAAAGUUACGUAUGAGCAUUUUGUGUAGGUCUAGUUUGCUAGCUGUUUU---UUGCAACAUUUUUUGCCUUUGUUGCAGACAAACUUU ((((((.((((((..((.(((.......))).))..))))))(.((((....)))))......---..((((((...........))))))))))))..... ( -24.00) >consensus UUGUCUGCUGCACUUAAAGUUACGUAUGAGCAUUUUGUGUAGGUCUAGUUUGCCAGCUUU_UU___UUGCAAC______UGCCUUUGUUGCAGACAAACUUU (((((((((((((..((.(((.......))).))..)))))..............((...........))...................))))))))..... (-15.16 = -15.42 + 0.25)

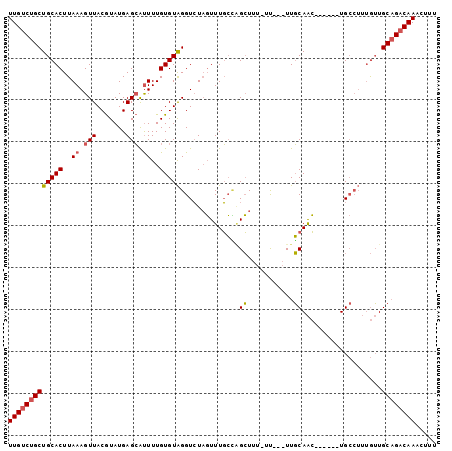
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:18:20 2006