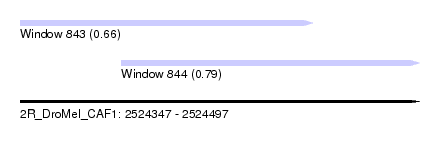
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 2,524,347 – 2,524,497 |
| Length | 150 |
| Max. P | 0.794549 |

| Location | 2,524,347 – 2,524,457 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 81.56 |
| Mean single sequence MFE | -36.35 |
| Consensus MFE | -21.40 |
| Energy contribution | -20.90 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.660364 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2524347 110 + 20766785 GAAAACAAAAGACUGCGUAGUACGUGUGGAAAAGUGUUGUGCAAGGGA--------GUCGGGCGGCGCGUAUGCUGCGCUUCCUCUUCUCCUUGACAAAGUGCAGUGAACUUUUCCCA ..............(((.....)))(.((((((((.(((..(.(((((--------(..(((.((((((.....)))))).)))...))))))......)..)))...))))))))). ( -35.70) >DroSim_CAF1 2457 118 + 1 GAAAACAAAAGACUGCGUAGUACGUGUGGAAAAGUGUUGUGCAAGGAGUGCGGGGCGGAGGGUGAAGCGUAUGCUGCGCUUCCUCUUCUCCUUGACAGAGUGCAGUGAACUUUUCCCA ..............(((.....)))(.((((((((....((((....((.(((((.((((((.(((((((.....))))))))))))).)))))))....))))....))))))))). ( -43.30) >DroEre_CAF1 4830 102 + 1 GAAAACAAAAGACUGCGCAGUAUGCGAAGAAAAGUGGG------GGCA--------AUGGG--AAAGCGUAUGCUGCGCUUCCUCUUCUCCUUGACAAAGUGCAGUGAACUUUUCCCA .....((.....((.(((.....))).)).....))..------....--------.((((--((((.((.((((((((((..((........))..)))))))))).)))))))))) ( -30.40) >DroYak_CAF1 2506 104 + 1 GAAAAUAAAAGACUGCGUAGUACGUGAUGAAAAGUACG------GGCA--------GUGGGGAAAAGCGUAUGCUGCGCUUCCUCUUCUCCUUGACAAAGUGCAGUGAACUUUUCCCU ...........(((((...((((..........)))).------.)))--------))(((((((((....((((((((((..((........))..))))))))))..))))))))) ( -36.00) >consensus GAAAACAAAAGACUGCGUAGUACGUGAGGAAAAGUGUG______GGCA________GUGGGG_AAAGCGUAUGCUGCGCUUCCUCUUCUCCUUGACAAAGUGCAGUGAACUUUUCCCA ..............(((.....)))..................................(((.((((.((.((((((((((..((........))..)))))))))).))))))))). (-21.40 = -20.90 + -0.50)

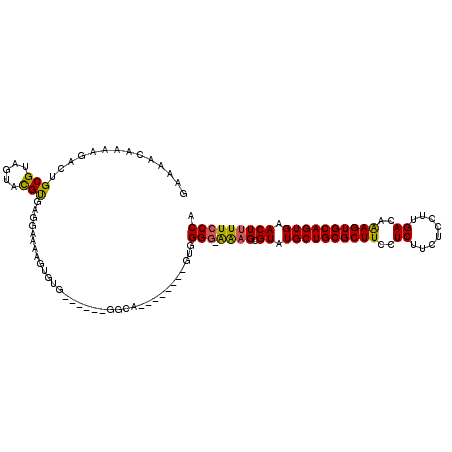
| Location | 2,524,385 – 2,524,497 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.74 |
| Mean single sequence MFE | -36.58 |
| Consensus MFE | -24.08 |
| Energy contribution | -24.57 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.794549 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2524385 112 + 20766785 GUGCAAGGGA--------GUCGGGCGGCGCGUAUGCUGCGCUUCCUCUUCUCCUUGACAAAGUGCAGUGAACUUUUCCCAUUUUCCCCACUUGAAUCUUCGAGUCCCAUUGACGCGGCCU ((((((((((--------.(((((..(...((.((((((((((..((........))..)))))))))).)).)..))).............((....)))).)))).))).)))..... ( -33.60) >DroSim_CAF1 2495 120 + 1 GUGCAAGGAGUGCGGGGCGGAGGGUGAAGCGUAUGCUGCGCUUCCUCUUCUCCUUGACAGAGUGCAGUGAACUUUUCCCAUUUUCCCCACUUGAAUCUUCGAGCCCCAUUGACGCGGCCU ((((((((.(.((((((..((.((.((((.((.((((((((((..((........))..)))))))))).)).)))))).))..))))..((((....))))))))).))).)))..... ( -41.70) >DroEre_CAF1 4868 103 + 1 ------GGCA--------AUGGG--AAAGCGUAUGCUGCGCUUCCUCUUCUCCUUGACAAAGUGCAGUGAACUUUUCCCAUU-UCCCCACUUGAAUCUUCGAGCCCCCUUCACGCGUCCU ------((.(--------(((((--((((.((.((((((((((..((........))..)))))))))).))))))))))))-.))...(((((....)))))................. ( -33.60) >DroYak_CAF1 2544 104 + 1 ------GGCA--------GUGGGGAAAAGCGUAUGCUGCGCUUCCUCUUCUCCUUGACAAAGUGCAGUGAACUUUUCCCUUU-U-CCCACUUGAAUCUUCGAGUCCCAUUGACGCGGCCU ------(((.--------(((((((((((....((((((((((..((........))..))))))))))..))))))))...-.-...((((((....))))))........))).))). ( -37.40) >consensus ______GGCA________GUGGGG_AAAGCGUAUGCUGCGCUUCCUCUUCUCCUUGACAAAGUGCAGUGAACUUUUCCCAUU_UCCCCACUUGAAUCUUCGAGCCCCAUUGACGCGGCCU .....................((((...((((..(((((((((..((........))..))))))))).....................(((((....)))))........)))).)))) (-24.08 = -24.57 + 0.50)

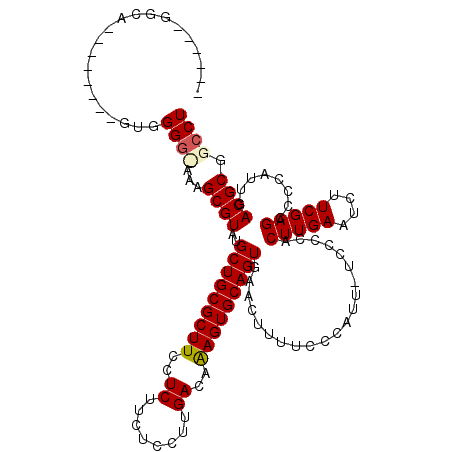
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:38:35 2006