| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,397,561 – 14,397,659 |
| Length | 98 |
| Max. P | 0.858845 |

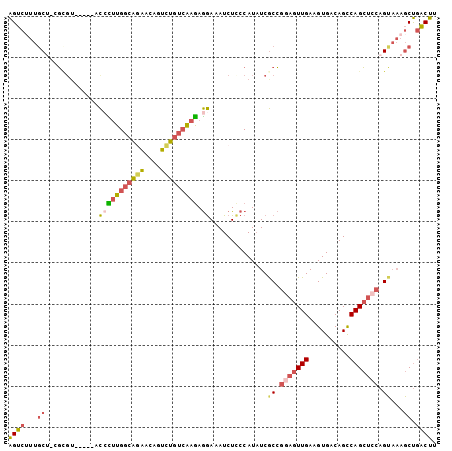
| Location | 14,397,561 – 14,397,659 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 77.67 |
| Mean single sequence MFE | -29.70 |
| Consensus MFE | -19.33 |
| Energy contribution | -20.11 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.748790 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14397561 98 + 20766785 AAGUCAGCUUUACUGGAGCUGACUGUCACUUCAACUCCGGCGAUAUGGGAGAUUUCCUCUUGACAGACUGUUCUGCCAAGGGU-----ACGCC-AGCAAAGACU .(((((((((.....)))))))))(((.......((((.(.....).)))).....((((((.((((....)))).)))))).-----.....-......))). ( -31.60) >DroSec_CAF1 29725 98 + 1 AAGUCAGCUUUGCUGGAGCUGGCUGUCACUUCAACUCCGGCGAAAUGGAAGAUUUCCUCUUGACAGAUUGUUCUGCCAAAGGU-----ACGCA-GGCAAAGACU ..(((.((((((((((((.(((........))).))))))))))...........(((.(((.((((....)))).)))))).-----..)).-)))....... ( -28.80) >DroSim_CAF1 33084 98 + 1 AAGUCAGCUUUACUGGAGCUGGCUGUCACUUCAACUGCGGCGAUAUGGGAGAUUUCCUCUUGACAGACUGUUCUGCCAAGGGU-----ACGCC-AGCAAAGACU .(((((((((.....)))))))))...........(((((((.((((((......)))((((.((((....)))).)))).))-----)))))-.)))...... ( -32.50) >DroEre_CAF1 31491 95 + 1 AAGUCAGCUUUACUGGAGCUGGCUGUCACUUCAACUCCGGCGAUAUGGGAGAUUUCCUCUUGACAGACUGCUAUGCCAUAUG---------CGAAGCAAAGACU .(((((((((.....)))))))))(((.......((((.(.....).)))).................((((.(((.....)---------)).))))..))). ( -26.90) >DroYak_CAF1 30908 98 + 1 AAGUCAGCUUUACUGGAGCUGGCUGUCACUUCAACUCCGGCGAUAUGGGAGAUUUCCUCUUGACAGACUGCUCUGCCAAGGGA-----ACACG-AGCAACGACU .(((((((((.....)))))))))(((.......((((.(.....).))))..((((.((((.((((....)))).)))))))-----)....-......))). ( -32.90) >DroPer_CAF1 30468 94 + 1 -AGCUGG-----CUG--CCUGGCUGUCACUUCAACAUUUGUGAUAUGGCAGAUGUUUUUUCCCCAAAU-GUCUCCCCAAAUACCCUUAACGAA-GGGACAGGCC -......-----..(--((((..((((((..........)))))).((.((((((((.......))))-)))).))......(((((....))-))).))))). ( -25.50) >consensus AAGUCAGCUUUACUGGAGCUGGCUGUCACUUCAACUCCGGCGAUAUGGGAGAUUUCCUCUUGACAGACUGUUCUGCCAAAGGU_____ACGCA_AGCAAAGACU .(((((((((.....)))))))))(((.((((...............)))).....((((((.((((....)))).))))))..................))). (-19.33 = -20.11 + 0.78)


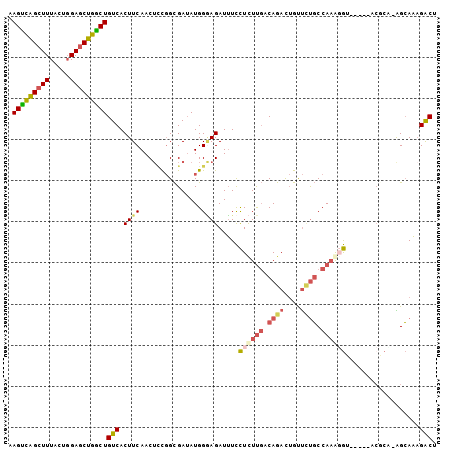
| Location | 14,397,561 – 14,397,659 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 77.67 |
| Mean single sequence MFE | -31.35 |
| Consensus MFE | -18.79 |
| Energy contribution | -19.63 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.858845 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14397561 98 - 20766785 AGUCUUUGCU-GGCGU-----ACCCUUGGCAGAACAGUCUGUCAAGAGGAAAUCUCCCAUAUCGCCGGAGUUGAAGUGACAGUCAGCUCCAGUAAAGCUGACUU ((.(((((((-((((.-----...(((((((((....))))))))).(((....))).....))))((((((((........)))))))))))))))))..... ( -39.70) >DroSec_CAF1 29725 98 - 1 AGUCUUUGCC-UGCGU-----ACCUUUGGCAGAACAAUCUGUCAAGAGGAAAUCUUCCAUUUCGCCGGAGUUGAAGUGACAGCCAGCUCCAGCAAAGCUGACUU ((.((((((.-.(((.-----...(((((((((....))))))))).(((.....)))....))).(((((((..........))))))).))))))))..... ( -32.00) >DroSim_CAF1 33084 98 - 1 AGUCUUUGCU-GGCGU-----ACCCUUGGCAGAACAGUCUGUCAAGAGGAAAUCUCCCAUAUCGCCGCAGUUGAAGUGACAGCCAGCUCCAGUAAAGCUGACUU ((.(((((((-((.((-----.(((((((((((....))))))))).))............((((..(....)..))))......)).)))))))))))..... ( -32.10) >DroEre_CAF1 31491 95 - 1 AGUCUUUGCUUCG---------CAUAUGGCAUAGCAGUCUGUCAAGAGGAAAUCUCCCAUAUCGCCGGAGUUGAAGUGACAGCCAGCUCCAGUAAAGCUGACUU ((((...((((..---------(....(((.......(((....)))(((....)))......)))(((((((..........))))))).)..)))).)))). ( -28.40) >DroYak_CAF1 30908 98 - 1 AGUCGUUGCU-CGUGU-----UCCCUUGGCAGAGCAGUCUGUCAAGAGGAAAUCUCCCAUAUCGCCGGAGUUGAAGUGACAGCCAGCUCCAGUAAAGCUGACUU (((((..(((-....(-----((((((((((((....))))))))).))))............((.(((((((..........))))))).))..)))))))). ( -32.70) >DroPer_CAF1 30468 94 - 1 GGCCUGUCCC-UUCGUUAAGGGUAUUUGGGGAGAC-AUUUGGGGAAAAAACAUCUGCCAUAUCACAAAUGUUGAAGUGACAGCCAGG--CAG-----CCAGCU- (((.((((.(-((((......((.....((.(((.-.(((.......)))..))).)).....))......))))).))))))).((--(..-----...)))- ( -23.20) >consensus AGUCUUUGCU_CGCGU_____ACCCUUGGCAGAACAGUCUGUCAAGAGGAAAUCUCCCAUAUCGCCGGAGUUGAAGUGACAGCCAGCUCCAGUAAAGCUGACUU ((((...((.............(((((((((((....))))))))).))..............((.(((((((..........))))))).))...)).)))). (-18.79 = -19.63 + 0.84)

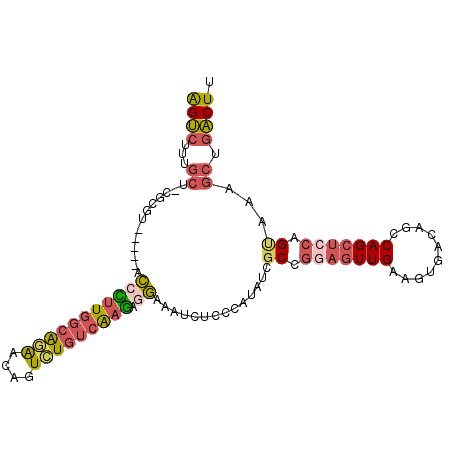
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:18:14 2006