
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,387,684 – 14,387,775 |
| Length | 91 |
| Max. P | 0.924275 |

| Location | 14,387,684 – 14,387,775 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 83.23 |
| Mean single sequence MFE | -29.77 |
| Consensus MFE | -16.52 |
| Energy contribution | -16.27 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.743651 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
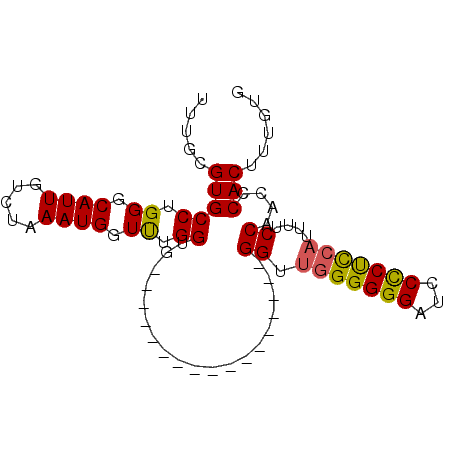
>2R_DroMel_CAF1 14387684 91 + 20766785 UUUGCGUGCCUGGGCAUUGUCUAAAUGGUUUGGUGUGAGGAGGGGGGGUGGGGGGUGGGGGGAUCCUCUCCAUUUUCCAACCCACUUUGUG ....((..((..(.((((.....)))).)..))..))(.((((.(((.(((..(((((((((...)))))))))..))).))).)))).). ( -38.00) >DroSec_CAF1 19864 74 + 1 UUUGCGUGCCUGGGCAUUGUCUAAAUGGUUUGGUG-----------------GGUUGGGGGGAUCCCCUCCAUUUUCCAACCCACUUUGUG .....((((....))))..............((((-----------------(((((((((....)))........))))))))))..... ( -24.60) >DroSim_CAF1 23121 74 + 1 UUUGCGUGCCUGGGCAUUGUCUAAAUGCUUUGGUG-----------------GGUUGGGGGGAUCCCCUCCAUUUUCCAACCCACUUUGUG .......(((..((((((.....))))))..)))(-----------------(((((((((....)))........)))))))........ ( -27.40) >DroEre_CAF1 21635 72 + 1 UUUGCGUGCCUGGGCAUUGUCUAAAUGGUCUGGGG-----------------GG--AGGGGGAUCCCCCUCAUUUUCCAACCCACUUGGUG .....((((....))))...((((.(((..(((..-----------------((--(((((....))))))..)..)))..))).)))).. ( -29.10) >consensus UUUGCGUGCCUGGGCAUUGUCUAAAUGGUUUGGUG_________________GGUUGGGGGGAUCCCCUCCAUUUUCCAACCCACUUUGUG .....(((((.((.((((.....)))).)).))...................((.(((((((...)))))))....))....)))...... (-16.52 = -16.27 + -0.25)

| Location | 14,387,684 – 14,387,775 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 83.23 |
| Mean single sequence MFE | -22.75 |
| Consensus MFE | -13.56 |
| Energy contribution | -13.62 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.924275 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14387684 91 - 20766785 CACAAAGUGGGUUGGAAAAUGGAGAGGAUCCCCCCACCCCCCACCCCCCCUCCUCACACCAAACCAUUUAGACAAUGCCCAGGCACGCAAA ....((((((.((((.....((((.((...................)).)))).....)))).))))))......(((....)))...... ( -20.41) >DroSec_CAF1 19864 74 - 1 CACAAAGUGGGUUGGAAAAUGGAGGGGAUCCCCCCAACC-----------------CACCAAACCAUUUAGACAAUGCCCAGGCACGCAAA ......(((((((((........(((....)))))))))-----------------)))................(((....)))...... ( -22.70) >DroSim_CAF1 23121 74 - 1 CACAAAGUGGGUUGGAAAAUGGAGGGGAUCCCCCCAACC-----------------CACCAAAGCAUUUAGACAAUGCCCAGGCACGCAAA ......(((((((((........(((....)))))))))-----------------)))................(((....)))...... ( -22.70) >DroEre_CAF1 21635 72 - 1 CACCAAGUGGGUUGGAAAAUGAGGGGGAUCCCCCU--CC-----------------CCCCAGACCAUUUAGACAAUGCCCAGGCACGCAAA ....((((((.((((.....((((((....)))))--).-----------------..)))).))))))......(((....)))...... ( -25.20) >consensus CACAAAGUGGGUUGGAAAAUGGAGGGGAUCCCCCCAACC_________________CACCAAACCAUUUAGACAAUGCCCAGGCACGCAAA ......((((.((((........((((....)))).............................((((.....)))).)))).).)))... (-13.56 = -13.62 + 0.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:18:10 2006