| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,375,053 – 14,375,184 |
| Length | 131 |
| Max. P | 0.998881 |

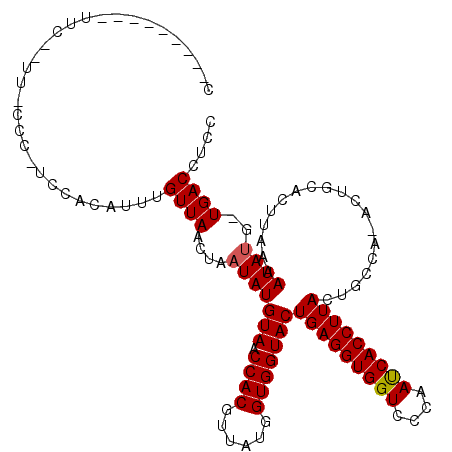
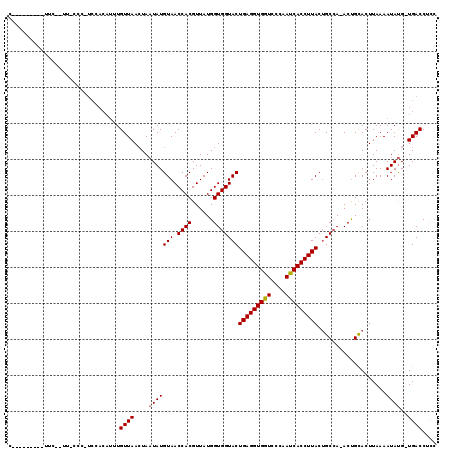
| Location | 14,375,053 – 14,375,163 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 75.14 |
| Mean single sequence MFE | -22.70 |
| Consensus MFE | -18.69 |
| Energy contribution | -18.80 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.82 |
| SVM decision value | 3.27 |
| SVM RNA-class probability | 0.998881 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14375053 110 + 20766785 C---------UUCUAUUUCCCUUCCACAUUUGUUAAAUAUUAUGUAACCACGUUAUGGUGGUACUGAGGUGGUCCCAACCACCUUACUGCAAAACCGUACUUAAAAUACGUUGACCUUU .---------.....................(((((.((((((((....))))((((((.(((.(((((((((....))))))))).)))...)))))).....))))..))))).... ( -24.30) >DroSec_CAF1 7517 117 + 1 CUUCCCCUCCUUCCCUUGCCCAUCCACAUUUGUUAACUAAUAUGUAACCACGUUAUGGUGGUACUGAGGUGGUCCCAAUCACCUUACAGCCA-ACUGCACUUAAAAUAUG-UGACCUCC ........................(((((...((((.......(((.((((......)))))))(((((((((....)))))))))(((...-.)))...))))...)))-))...... ( -22.60) >DroSim_CAF1 7541 88 + 1 C-----------------------------UGUUAACUAAUAUGUAACCACGUUAUGGUGGUACUGAGGUGGUCCCAAUCACCUUACUGCCA-ACUGCACUUAAAAUAUG-UGACCUCC .-----------------------------.........((((((.((((.....))))((((.(((((((((....))))))))).)))).-............)))))-)....... ( -21.20) >consensus C_________UUC__UU_CCC_UCCACAUUUGUUAACUAAUAUGUAACCACGUUAUGGUGGUACUGAGGUGGUCCCAAUCACCUUACUGCCA_ACUGCACUUAAAAUAUG_UGACCUCC ...............................((((....(((((((.((((......)))))))(((((((((....)))))))))...................))))..)))).... (-18.69 = -18.80 + 0.11)

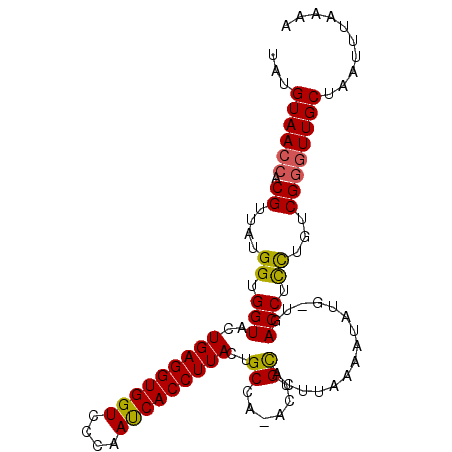
| Location | 14,375,084 – 14,375,184 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 90.60 |
| Mean single sequence MFE | -27.78 |
| Consensus MFE | -22.85 |
| Energy contribution | -22.63 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.847059 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14375084 100 + 20766785 UAUGUAACCACGUUAUGGUGGUACUGAGGUGGUCCCAACCACCUUACUGCAAAACCGUACUUAAAAUACGUUGACCUUUUGUCGCCUUGCCAAUUUAAUA .(((((.......((((((.(((.(((((((((....))))))))).)))...)))))).......))))).(((.....)))................. ( -25.64) >DroSec_CAF1 7557 98 + 1 UAUGUAACCACGUUAUGGUGGUACUGAGGUGGUCCCAAUCACCUUACAGCCA-ACUGCACUUAAAAUAUG-UGACCUCCUGUCGGGUUGCUAAUUUAAAA ...((((((.((....((.(((..(((((((((....)))))))))(((...-.)))(((.........)-))))).))...)))))))).......... ( -27.80) >DroSim_CAF1 7552 98 + 1 UAUGUAACCACGUUAUGGUGGUACUGAGGUGGUCCCAAUCACCUUACUGCCA-ACUGCACUUAAAAUAUG-UGACCUCCUGUCGGGUUGCUAAUUUAAAA ...((((((.((....(((((((.(((((((((....))))))))).)))).-....(((.........)-)))))......)))))))).......... ( -29.90) >consensus UAUGUAACCACGUUAUGGUGGUACUGAGGUGGUCCCAAUCACCUUACUGCCA_ACUGCACUUAAAAUAUG_UGACCUCCUGUCGGGUUGCUAAUUUAAAA ...((((((.((....((.(((..(((((((((....)))))))))..((......))...............))).))...)))))))).......... (-22.85 = -22.63 + -0.21)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:18:06 2006