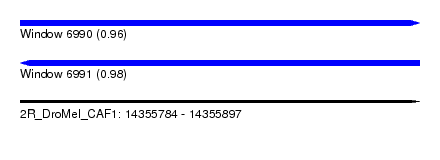
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,355,784 – 14,355,897 |
| Length | 113 |
| Max. P | 0.983371 |

| Location | 14,355,784 – 14,355,897 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 77.10 |
| Mean single sequence MFE | -38.78 |
| Consensus MFE | -22.07 |
| Energy contribution | -21.99 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.57 |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.964334 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
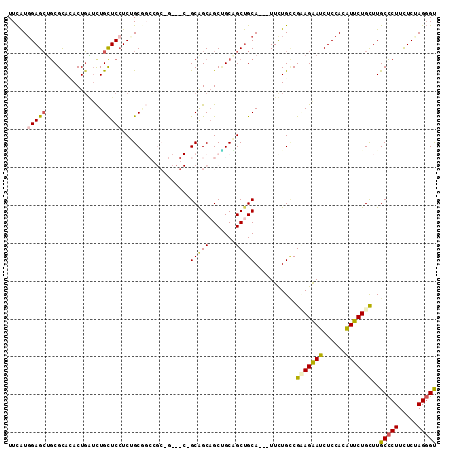
>2R_DroMel_CAF1 14355784 113 + 20766785 UUCAUGGAGCUGCGCACACUGAUCUGCUCCUCUGCUGCCGCAGCCGCUGCAGCUGCAGCAGCUGCAUUAUUUUGCCGCAGAAUCUCCACAUUCUGUUUGCCCUUUUCUAGGGU ....((((((((((((.........((......))....(((((.(((((....))))).))))).......)).)))))...)))))..........(((((.....))))) ( -41.40) >DroPse_CAF1 8677 98 + 1 UUCAUGGAGCUGCGCAGACUGGUUUGCUCCUCCGCG------------GCAGCAGCCGCCGCUGCA---UUCUGUCGAAGGAUCUCCACGUUCUGCUUGCCCUUCUCCAGGGC ....(((((..(((((((((((...(.((((.((((------------(..(((((....))))).---..))).)).)))).).))).).)))))..))....))))).... ( -34.00) >DroGri_CAF1 11974 107 + 1 UUUGUGGAGGAACGUAUACUCUGCUGCUCCUCGGCAGCGGCAA---CGGCAGCAGCUGCUGCCGCA---UUCUGACGCAGAAUCUCCACAUUCUGUUUGCUCUUCUCAAGCGU ..((((((((..........((((((((....))))))))...---(((((((....)))))))..---(((((...)))))))))))))....(((((.......))))).. ( -41.60) >DroEre_CAF1 8813 113 + 1 UUCAUGGAGCUGCGCACACUGAUCUGCUCAUCUGCGGCCGCCGCCGCUGCGGCAGCUGCAGCUGCAUUAUUUUGCCGUAGAAUCUCCACAUUCUGCUUGCCCUUUUCUAGGGU ....((((((((((((.(.(((..(((....(((((((.(((((....))))).)))))))..)))))).).)).)))))...)))))..........(((((.....))))) ( -42.90) >DroYak_CAF1 8728 113 + 1 UUCAUUGAGCUGCGUACACUGAUCUGCUCCUCUGCUGCCGCCGCCGCCGCAGCAGCUGCAGCUGCAUUAUUUUGCCGUAGAAUCUCCACAUUCUGCUUGCCCUUUUCUAGGGU .......(((((((((........)))....(((((((.((....)).)))))))..))))))(((......))).(((((((......)))))))..(((((.....))))) ( -38.70) >DroPer_CAF1 8816 98 + 1 UUCAUGGAGCUGCGCAGACUGGUUUGUUCCUCCGCG------------GCAGCAGCCGCCGCUGCA---UUCUGUCGAAGGAUCUCCACGUUCUGCUUGCCCUUCUCCAGGGC ....(((((..(((((((((((...(.((((.((((------------(..(((((....))))).---..))).)).)))).).))).).)))))..))....))))).... ( -34.10) >consensus UUCAUGGAGCUGCGCACACUGAUCUGCUCCUCUGCGGCCGC_G___C_GCAGCAGCUGCAGCUGCA___UUCUGCCGAAGAAUCUCCACAUUCUGCUUGCCCUUCUCUAGGGU .....(((((...............)))))..................(((((.......)))))...........(((((((......)))))))..(((((.....))))) (-22.07 = -21.99 + -0.08)

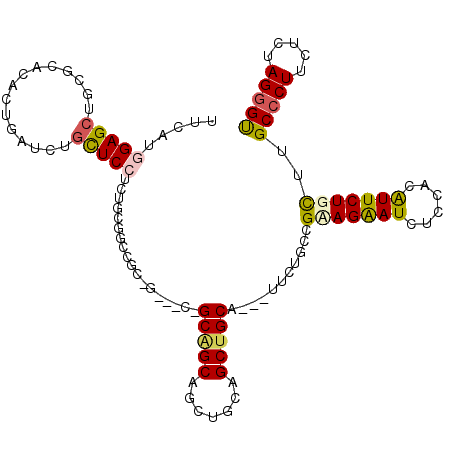
| Location | 14,355,784 – 14,355,897 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 77.10 |
| Mean single sequence MFE | -42.27 |
| Consensus MFE | -21.20 |
| Energy contribution | -21.23 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.50 |
| SVM decision value | 1.94 |
| SVM RNA-class probability | 0.983371 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14355784 113 - 20766785 ACCCUAGAAAAGGGCAAACAGAAUGUGGAGAUUCUGCGGCAAAAUAAUGCAGCUGCUGCAGCUGCAGCGGCUGCGGCAGCAGAGGAGCAGAUCAGUGUGCGCAGCUCCAUGAA .((((.....))))..........((((((.((((((.((.......((((((((((((....)))))))))))))).))))))..(((.(....).)))....))))))... ( -47.91) >DroPse_CAF1 8677 98 - 1 GCCCUGGAGAAGGGCAAGCAGAACGUGGAGAUCCUUCGACAGAA---UGCAGCGGCGGCUGCUGC------------CGCGGAGGAGCAAACCAGUCUGCGCAGCUCCAUGAA (((((.....)))))........(((((((.(((((((.(.(..---.(((((....)))))..)------------.))))))))(((........)))....))))))).. ( -42.20) >DroGri_CAF1 11974 107 - 1 ACGCUUGAGAAGAGCAAACAGAAUGUGGAGAUUCUGCGUCAGAA---UGCGGCAGCAGCUGCUGCCG---UUGCCGCUGCCGAGGAGCAGCAGAGUAUACGUUCCUCCACAAA ..((((.....))))........((((((((((((((.......---.((((((((.((....)).)---)))))))(((......))))))))))........))))))).. ( -42.70) >DroEre_CAF1 8813 113 - 1 ACCCUAGAAAAGGGCAAGCAGAAUGUGGAGAUUCUACGGCAAAAUAAUGCAGCUGCAGCUGCCGCAGCGGCGGCGGCCGCAGAUGAGCAGAUCAGUGUGCGCAGCUCCAUGAA .((((.....))))...((....(((((.....)))))))......(((.((((((.((((((((....)))))))).(((.((((.....))).).))))))))).)))... ( -43.40) >DroYak_CAF1 8728 113 - 1 ACCCUAGAAAAGGGCAAGCAGAAUGUGGAGAUUCUACGGCAAAAUAAUGCAGCUGCAGCUGCUGCGGCGGCGGCGGCAGCAGAGGAGCAGAUCAGUGUACGCAGCUCAAUGAA .((((.....)))).........(((((.....)))))(((......)))((((((..(((((((.((....)).)))))))..(.(((......))).)))))))....... ( -38.10) >DroPer_CAF1 8816 98 - 1 GCCCUGGAGAAGGGCAAGCAGAACGUGGAGAUCCUUCGACAGAA---UGCAGCGGCGGCUGCUGC------------CGCGGAGGAACAAACCAGUCUGCGCAGCUCCAUGAA (((((.....)))))........(((((((...((.((.((((.---....(((((((...))))------------)))...((......))..)))))).))))))))).. ( -39.30) >consensus ACCCUAGAAAAGGGCAAGCAGAAUGUGGAGAUUCUACGGCAAAA___UGCAGCUGCAGCUGCUGC_G___C_GCGGCCGCAGAGGAGCAGAUCAGUGUGCGCAGCUCCAUGAA .((((.....)))).........(((((((.((((.(...........(((((....))))).((.((.......)).)).)))))((......))........))))))).. (-21.20 = -21.23 + 0.03)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:17:59 2006