
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,334,678 – 14,334,776 |
| Length | 98 |
| Max. P | 0.572716 |

| Location | 14,334,678 – 14,334,776 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 74.53 |
| Mean single sequence MFE | -29.42 |
| Consensus MFE | -13.75 |
| Energy contribution | -14.12 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.572716 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
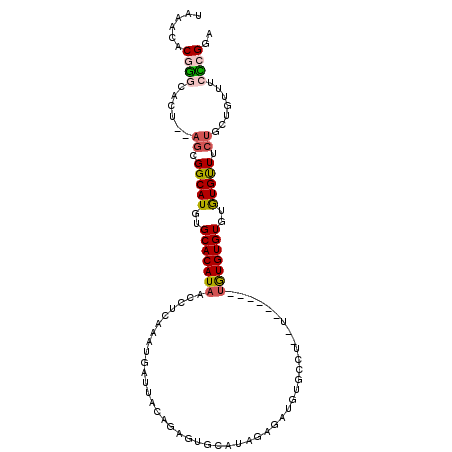
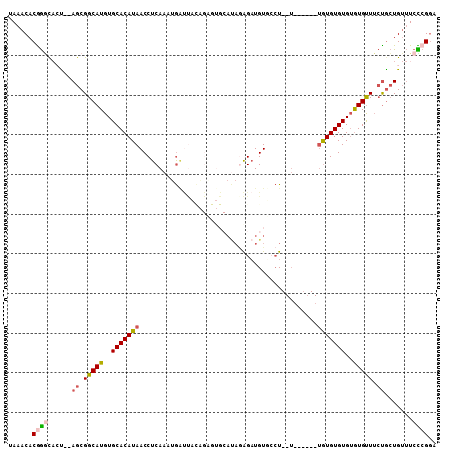
>2R_DroMel_CAF1 14334678 98 + 20766785 UAAACACGGGCAAU--AGCGGCAUGUGCACAUAACCUCAAAUGAUUACAGAGUGCAUAGAGAUGUUGCU--U------UGUGUGUGUGUGUUUCUGCUGUUUCCCGGA ......((((.(((--(((((...(..((((((...((....)).((((((((((((....))).))))--)------))))))))))..)..)))))))).)))).. ( -35.40) >DroVir_CAF1 562 103 + 1 AAAACACGACUUACGUAGCGGCAUGUGCACAUAACCUCAAAAAGCCUUUGUAUGC----GGCUGUGCGUGUUUGUGUGUGUGUGUGGGUGCUAGCGGUG-CUAUGGAA ....(((..((...(((.(.(((((..(((((((((.((...((((.........----)))).)).).).)))))))..))))).).))).))..)))-........ ( -30.00) >DroSec_CAF1 188 98 + 1 UAAACACGGGCACU--AGCGGCAUGUGCACAUAACCUCAAAUGAUUACAGAGUGCAUAAAGAUGUUCCU--U------UGUGUGUGUGUGUUUCUACUGUUUCCCGGA ......((((((.(--((..(((((..(((......((....))..((((((.((((....))))..))--)------))))))..)))))..))).))...)))).. ( -27.60) >DroSim_CAF1 188 98 + 1 UAAACACGGGCACU--AGCGGCAUGUGCACAUAACCUCAAAUGAUUACAGAGUGCAUAGAGAUGUUGCU--U------UAUGUGUGUGUGUUUCUGCUGUUUCCCGGA ......((((....--(((((((...(((((((..(((...........))).((((((((......))--)------))))).)))))))...))))))).)))).. ( -31.70) >DroEre_CAF1 187 85 + 1 UAAACACAGACUCC--AGCGGCAUGUGCACAUAACCUCAAAUGC-----------AUAGAGAUGUGCCU----------GUGUGUGUGUGUUUCUGCUGUUUCUCGGA ...........(((--(((((((...(((((((....((...((-----------(((....)))))..----------.))..)))))))...)))))))....))) ( -23.90) >DroYak_CAF1 188 93 + 1 UAAACACGGGCACU--AGCGGCAUGUGCACAUAACCUCAAAUGAUU---------ACAGAGAUGUGCCU--UUGUU--UGUGUGUGUAUGUUUCUGCUGUUUCUCGGA ......((((...(--(((((((((..(((((((..((....))..---------((((((......))--)))))--))))))..))))...))))))...)))).. ( -27.90) >consensus UAAACACGGGCACU__AGCGGCAUGUGCACAUAACCUCAAAUGAUUACAGAGUGCAUAGAGAUGUGCCU__U______UGUGUGUGUGUGUUUCUGCUGUUUCCCGGA ......((((......((.(((((..(((((((.............................................)))))))..))))).)).......)))).. (-13.75 = -14.12 + 0.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:17:52 2006