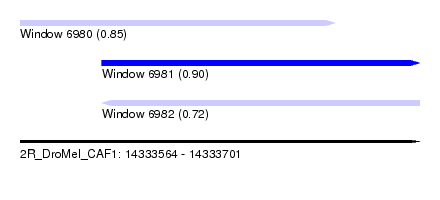
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,333,564 – 14,333,701 |
| Length | 137 |
| Max. P | 0.902294 |

| Location | 14,333,564 – 14,333,672 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.23 |
| Mean single sequence MFE | -24.35 |
| Consensus MFE | -19.10 |
| Energy contribution | -19.52 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.846742 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
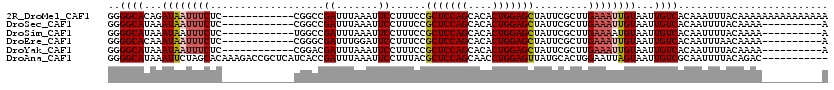
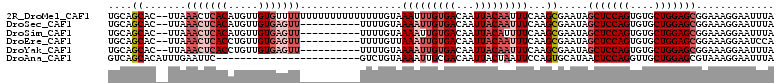
>2R_DroMel_CAF1 14333564 108 + 20766785 GGGGCACAGAUAAUUUCUC------------CGGCCGAUUUAAAUUCCUUUCCGCUCCAGCACACUGGAGCUAUUCGCUUGAAAUUGUAAUUGUCACAAAUUUACAAAAAAAAAAAAAAA ..((((...((((((((..------------.(((.((.......))......(((((((....))))))).....))).))))))))...))))......................... ( -24.50) >DroSec_CAF1 9772 98 + 1 GGGGCAUAAAUAAUUUCUC------------CGGCCGAUUUAAAUUCCUUUCCGCUCCAGCACACUGGAGCUAUUCGCUUGAAAUUGUAAUUGUCACAAUUUUACAAAA----------A ..((((...((((((((..------------.(((.((.......))......(((((((....))))))).....))).))))))))...))))..............----------. ( -24.10) >DroSim_CAF1 10509 98 + 1 GGGGCAUAAAUAAUUUCUC------------UGGCCGAUUUAAAUUCCUUUCCGCUCCAGCACACUGGAGCUAUUCGCUUGAAAAUGUAAUUGUCACAAUUUUACAAAA----------A ..((((...(((.((((..------------.(((.((.......))......(((((((....))))))).....))).)))).)))...))))..............----------. ( -21.10) >DroEre_CAF1 9783 98 + 1 GGGGCACAAAUAAUUUCUC------------CGGGCGAUUUGGAUUCCUUUCCGCUCCAGCACACUGGAGCUAUUCGCUUGAAAUUGUAAUUGUCACAAUUUAACAAAA----------A ..((((...((((((((..------------.((((((...(((......)))(((((((....)))))))...))))))))))))))...))))..............----------. ( -31.30) >DroYak_CAF1 10268 98 + 1 GGGGCAUAAAUAAUUUCUC------------CGGACGAUUUAAAUUCCUUUCCGCUCCAGCACACUGGAGCUAUUCGCUUGAAAUUGUAAUUGUCACAAUUUUACAAAA----------A ..((((...((((((((..------------.(((.((.......))...)))(((((((....))))))).........))))))))...))))..............----------. ( -24.60) >DroAna_CAF1 10845 109 + 1 GGGGCAUAAAUUCUAGCACAAAGACCGCUCAUCACCGAUUUAAAUUCCUUUACGCUCCAGCAACCUGGAGUUAUGCACUGGAAUUAGUAAUUGUCGCAAUUUUACAGAC----------- (.((((..((((((((..((..((....)).......................(((((((....)))))))..))..))))))))......)))).)............----------- ( -20.50) >consensus GGGGCAUAAAUAAUUUCUC____________CGGCCGAUUUAAAUUCCUUUCCGCUCCAGCACACUGGAGCUAUUCGCUUGAAAUUGUAAUUGUCACAAUUUUACAAAA__________A ..((((...((((((((...................((.......))......(((((((....))))))).........))))))))...))))......................... (-19.10 = -19.52 + 0.42)
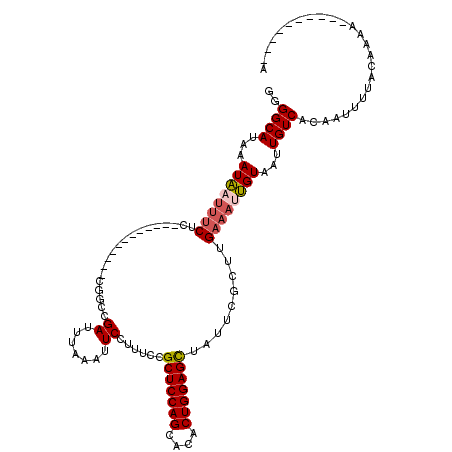
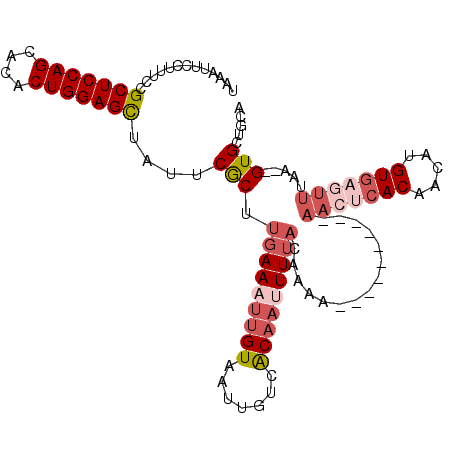
| Location | 14,333,592 – 14,333,701 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 83.88 |
| Mean single sequence MFE | -26.40 |
| Consensus MFE | -18.25 |
| Energy contribution | -20.08 |
| Covariance contribution | 1.84 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.74 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.902294 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14333592 109 + 20766785 UAAAUUCCUUUCCGCUCCAGCACACUGGAGCUAUUCGCUUGAAAUUGUAAUUGUCACAAAUUUACAAAAAAAAAAAAAAAAAACACAACAUGUGAGUUUAA--GUGCUGCA .............(((((((....)))))))....((((((((.((((((...........))))))................(((.....)))..)))))--)))..... ( -23.50) >DroSec_CAF1 9800 99 + 1 UAAAUUCCUUUCCGCUCCAGCACACUGGAGCUAUUCGCUUGAAAUUGUAAUUGUCACAAUUUUACAAAA----------AACUCACAACAUGUGAGUUUAA--GUGCUGCA .............(((((((....)))))))....(((((((((((((.......)))))))).....(----------(((((((.....))))))))))--)))..... ( -29.30) >DroSim_CAF1 10537 99 + 1 UAAAUUCCUUUCCGCUCCAGCACACUGGAGCUAUUCGCUUGAAAAUGUAAUUGUCACAAUUUUACAAAA----------AACUCACAACAUGUGAGUUUAA--GUGCUGCA .............(((((((....)))))))....(((((((...(((((...........)))))...----------.((((((.....))))))))))--)))..... ( -26.70) >DroEre_CAF1 9811 99 + 1 UGGAUUCCUUUCCGCUCCAGCACACUGGAGCUAUUCGCUUGAAAUUGUAAUUGUCACAAUUUAACAAAA----------AACUCACAACAGGUGAGUUUAA--GUGCUGCA .(((......)))(((((((....)))))))....(((((.(((((((.......)))))))......(----------(((((((.....))))))))))--)))..... ( -30.90) >DroYak_CAF1 10296 99 + 1 UAAAUUCCUUUCCGCUCCAGCACACUGGAGCUAUUCGCUUGAAAUUGUAAUUGUCACAAUUUUACAAAA----------AACUCACAACAGGUGAGUUUAA--GUGCUGCA .............(((((((....)))))))....(((((((((((((.......)))))))).....(----------(((((((.....))))))))))--)))..... ( -28.90) >DroAna_CAF1 10885 87 + 1 UAAAUUCCUUUACGCUCCAGCAACCUGGAGUUAUGCACUGGAAUUAGUAAUUGUCGCAAUUUUACAGAC------------------------GAAUUCAAAUGUGCUGAC .............(((((((....)))))))...((((..((((......(((((...........)))------------------------))))))....)))).... ( -19.10) >consensus UAAAUUCCUUUCCGCUCCAGCACACUGGAGCUAUUCGCUUGAAAUUGUAAUUGUCACAAUUUUACAAAA__________AACUCACAACAUGUGAGUUUAA__GUGCUGCA .............(((((((....)))))))....(((.(((((((((.......)))))))))...............(((((((.....))))))).....)))..... (-18.25 = -20.08 + 1.84)

| Location | 14,333,592 – 14,333,701 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 83.88 |
| Mean single sequence MFE | -28.15 |
| Consensus MFE | -19.09 |
| Energy contribution | -21.20 |
| Covariance contribution | 2.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.724589 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
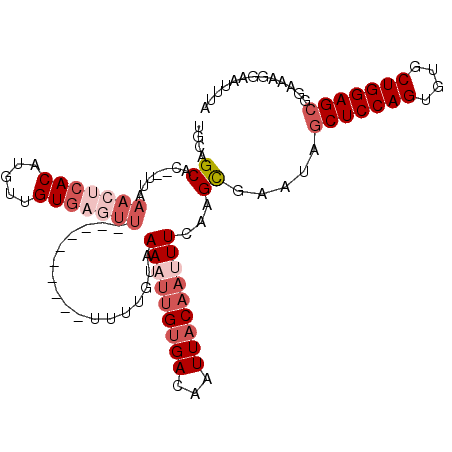
>2R_DroMel_CAF1 14333592 109 - 20766785 UGCAGCAC--UUAAACUCACAUGUUGUGUUUUUUUUUUUUUUUUUUGUAAAUUUGUGACAAUUACAAUUUCAAGCGAAUAGCUCCAGUGUGCUGGAGCGGAAAGGAAUUUA .((((((.--...........))))))((((((((((......(((((....((((((...))))))......)))))..(((((((....)))))))))))))))))... ( -24.40) >DroSec_CAF1 9800 99 - 1 UGCAGCAC--UUAAACUCACAUGUUGUGAGUU----------UUUUGUAAAAUUGUGACAAUUACAAUUUCAAGCGAAUAGCUCCAGUGUGCUGGAGCGGAAAGGAAUUUA .((.(((.--..((((((((.....)))))))----------)..))).(((((((((...)))))))))...)).....(((((((....)))))))............. ( -31.50) >DroSim_CAF1 10537 99 - 1 UGCAGCAC--UUAAACUCACAUGUUGUGAGUU----------UUUUGUAAAAUUGUGACAAUUACAUUUUCAAGCGAAUAGCUCCAGUGUGCUGGAGCGGAAAGGAAUUUA .((.(((.--..((((((((.....)))))))----------)..))).(((.(((((...))))).)))...)).....(((((((....)))))))............. ( -28.30) >DroEre_CAF1 9811 99 - 1 UGCAGCAC--UUAAACUCACCUGUUGUGAGUU----------UUUUGUUAAAUUGUGACAAUUACAAUUUCAAGCGAAUAGCUCCAGUGUGCUGGAGCGGAAAGGAAUCCA .((((((.--..((((((((.....)))))))----------)..))))(((((((((...)))))))))...)).....(((((((....)))))))(((......))). ( -33.60) >DroYak_CAF1 10296 99 - 1 UGCAGCAC--UUAAACUCACCUGUUGUGAGUU----------UUUUGUAAAAUUGUGACAAUUACAAUUUCAAGCGAAUAGCUCCAGUGUGCUGGAGCGGAAAGGAAUUUA .((.(((.--..((((((((.....)))))))----------)..))).(((((((((...)))))))))...)).....(((((((....)))))))............. ( -30.50) >DroAna_CAF1 10885 87 - 1 GUCAGCACAUUUGAAUUC------------------------GUCUGUAAAAUUGCGACAAUUACUAAUUCCAGUGCAUAACUCCAGGUUGCUGGAGCGUAAAGGAAUUUA (((.(((.((((......------------------------.......)))))))))).......((((((..(((....((((((....)))))).)))..)))))).. ( -20.62) >consensus UGCAGCAC__UUAAACUCACAUGUUGUGAGUU__________UUUUGUAAAAUUGUGACAAUUACAAUUUCAAGCGAAUAGCUCCAGUGUGCUGGAGCGGAAAGGAAUUUA ....((.......(((((((.....))))))).................(((((((((...)))))))))...)).....(((((((....)))))))............. (-19.09 = -21.20 + 2.11)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:17:50 2006