| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 2,517,547 – 2,517,647 |
| Length | 100 |
| Max. P | 0.623697 |

| Location | 2,517,547 – 2,517,647 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 51.95 |
| Mean single sequence MFE | -25.71 |
| Consensus MFE | -10.68 |
| Energy contribution | -9.80 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.42 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.623697 |
| Prediction | RNA |
| WARNING | Mean pairwise identity too low. |
Download alignment: ClustalW | MAF
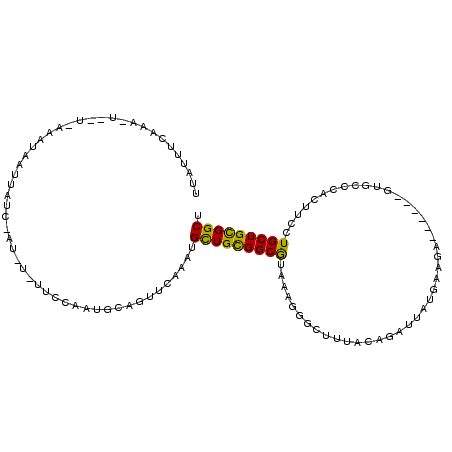
>2R_DroMel_CAF1 2517547 100 + 20766785 UUUUUUCCG--------GAUAUUUACGUAUCUAUUCCAUUGCAGUUCUCUUGCUGUGGCAACACGGGAUACCAACAGUAUGAAACCG-----UGCCCAGCUCCUGCUGCGGCU .......((--------(.....(((((...((((((...((((.....)))).(((....)))))))))...)).))).....)))-----.(((((((....)))).))). ( -29.60) >DroVir_CAF1 26869 102 + 1 UGAGAUCUAACUUUUCAUUUGAUUUUC------CUGCAAUUUAGUUCAAAUGUUGCGGCGUAAACAGCUUUACGGAUUAUACAGU-----UGUGCCCUCUUCCUGCUGUGGCU .((((.......))))((((((.....------............))))))((..(((((((((....)))))(((...(((...-----.)))......))).))))..)). ( -18.63) >DroGri_CAF1 26605 110 + 1 UUAUUUCAAAUUCAUAAAAUAAUCAUCAAUAU-UUUCAAUGCAGUUGGAAUGCUGCGGCGUAAAGGACUUUACCGAUUAUGGAGA--CAAAGUUCCGCUUUCGUGCUGCGGCU ..(((((((...(((((((((........)))-)))..)))...)))))))(((((((((((((....)))))(((...(((((.--.....)))))...))).)))))))). ( -28.90) >consensus UUAUUUCAAA_U__U_AAAUAAUUAUC_AU_U_UUCCAAUGCAGUUCAAAUGCUGCGGCGUAAAGGGCUUUACAGAUUAUGAAGA______GUGCCCACUUCCUGCUGCGGCU ...................................................(((((((((...........................................))))))))). (-10.68 = -9.80 + -0.88)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:38:32 2006