| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,327,978 – 14,328,068 |
| Length | 90 |
| Max. P | 0.714687 |

| Location | 14,327,978 – 14,328,068 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 96.28 |
| Mean single sequence MFE | -23.16 |
| Consensus MFE | -19.05 |
| Energy contribution | -19.13 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.672238 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
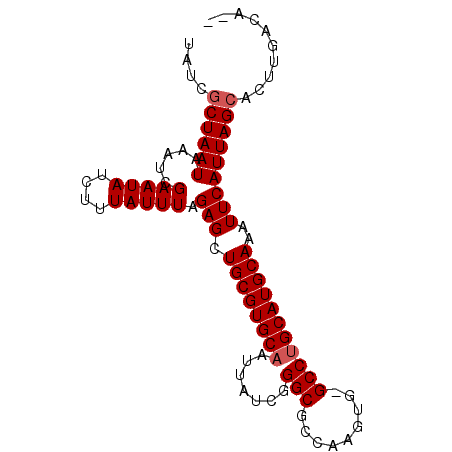
>2R_DroMel_CAF1 14327978 90 + 20766785 --UGUCAAGUGCUAAUGAAUUUGCAUGCCGGC-CACUUGGCGCCCGAUAAUUGCACGCAGCUCUAAAUAAAGAUAUUCGAUUUAUUAGCGUUA --........(((((((((((.((.(((..((-(....)))((.........))..)))))(((......))).....))))))))))).... ( -20.50) >DroSec_CAF1 4402 91 + 1 --UGUCAAGUGCUAAUGAAUUUGCAUGCAGGCGCACUUGGCGCCCGAUAAUUGCACGCAGCUCCAAAUAAAGAUAUUCGAUUUAUUAGCAAUA --.......(((((((((((((((.(((((((((.....))))).......)))).)))............(.....)))))))))))))... ( -26.41) >DroSim_CAF1 4534 91 + 1 --UGUCAAGUGCUAAUGAAUUUGCAUGCAGGCGCACUUGGCGCCCGAUAAUUGCACGCAGCUCUAAAUAAAGAUAUUCGAUUUAUUAGCGAUA --.......(((((((((((((((.(((((((((.....))))).......)))).)))..(((......))).....))))))))))))... ( -27.11) >DroEre_CAF1 4469 90 + 1 --UGUCAAGUGCUAAUGAAUUUGCAUGCAGGC-CACUUGGCGCCCGAUAAUUGCACGCAGCUCUAAAUAAAGAUAUUCGAUUUAUUAGUGAUA --........((((((((((((((.(((((((-(....))).........))))).)))..(((......))).....))))))))))).... ( -19.70) >DroYak_CAF1 4596 92 + 1 UUUGUCAAGUGCUAAUGAAUUUGCAUGCAGGC-CACUUGGCGCCCGAUAAUUGCACGCAGCUCUAAAUAAAGAUAUUCGAUUUAUUAGCGAUA .........(((((((((((((((.(((((((-(....))).........))))).)))..(((......))).....))))))))))))... ( -22.10) >consensus __UGUCAAGUGCUAAUGAAUUUGCAUGCAGGC_CACUUGGCGCCCGAUAAUUGCACGCAGCUCUAAAUAAAGAUAUUCGAUUUAUUAGCGAUA .........(((((((((((((((.(((((((.(.....).))).......)))).)))..(((......))).....))))))))))))... (-19.05 = -19.13 + 0.08)


| Location | 14,327,978 – 14,328,068 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 96.28 |
| Mean single sequence MFE | -24.34 |
| Consensus MFE | -18.75 |
| Energy contribution | -19.15 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.714687 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14327978 90 - 20766785 UAACGCUAAUAAAUCGAAUAUCUUUAUUUAGAGCUGCGUGCAAUUAUCGGGCGCCAAGUG-GCCGGCAUGCAAAUUCAUUAGCACUUGACA-- ....((((((.....(((((....))))).(((.(((((((......(((.(((...)))-.))))))))))..)))))))))........-- ( -23.80) >DroSec_CAF1 4402 91 - 1 UAUUGCUAAUAAAUCGAAUAUCUUUAUUUGGAGCUGCGUGCAAUUAUCGGGCGCCAAGUGCGCCUGCAUGCAAAUUCAUUAGCACUUGACA-- ...(((((((...(((((((....)))))))...((((((((.......(((((.....))))))))))))).....))))))).......-- ( -29.51) >DroSim_CAF1 4534 91 - 1 UAUCGCUAAUAAAUCGAAUAUCUUUAUUUAGAGCUGCGUGCAAUUAUCGGGCGCCAAGUGCGCCUGCAUGCAAAUUCAUUAGCACUUGACA-- ....((((((.....(((((....))))).(((.((((((((.......(((((.....)))))))))))))..)))))))))........-- ( -26.91) >DroEre_CAF1 4469 90 - 1 UAUCACUAAUAAAUCGAAUAUCUUUAUUUAGAGCUGCGUGCAAUUAUCGGGCGCCAAGUG-GCCUGCAUGCAAAUUCAUUAGCACUUGACA-- .....(((((.....(((((....))))).(((.((((((((......((.(((...)))-.))))))))))..)))))))).........-- ( -18.50) >DroYak_CAF1 4596 92 - 1 UAUCGCUAAUAAAUCGAAUAUCUUUAUUUAGAGCUGCGUGCAAUUAUCGGGCGCCAAGUG-GCCUGCAUGCAAAUUCAUUAGCACUUGACAAA ....((((((.....(((((....))))).(((.((((((((......((.(((...)))-.))))))))))..))))))))).......... ( -23.00) >consensus UAUCGCUAAUAAAUCGAAUAUCUUUAUUUAGAGCUGCGUGCAAUUAUCGGGCGCCAAGUG_GCCUGCAUGCAAAUUCAUUAGCACUUGACA__ ....((((((.....(((((....))))).(((.((((((((.......(((.........)))))))))))..))))))))).......... (-18.75 = -19.15 + 0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:17:46 2006