| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,325,074 – 14,325,176 |
| Length | 102 |
| Max. P | 0.511747 |

| Location | 14,325,074 – 14,325,176 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 76.97 |
| Mean single sequence MFE | -25.35 |
| Consensus MFE | -11.51 |
| Energy contribution | -13.32 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.45 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.511747 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
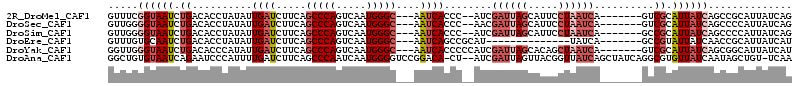
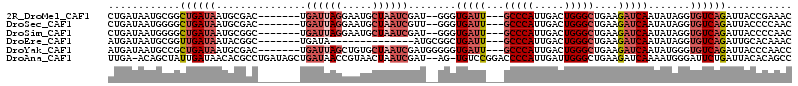
>2R_DroMel_CAF1 14325074 102 + 20766785 GUUUCGGUAAUCUGACACCUAUAUUGAUCUUCAGCCCAGUCAAUGGGC---AAUCACCC--AUCGAUUAGCAUUCCUAAUCA-------GUCGCAUUAUCAGCCGCAUUAUCAG .....((((((.((((.................(((((.....)))))---........--...((((((.....)))))).-------)))).)))))).............. ( -24.20) >DroSec_CAF1 1605 102 + 1 GUUGGGGUAAUCUGACACCUAUAUUGAUCUUCAGCCCAGUCAAUGGGC---AAUCACCC--AACGAUUAGCAUUCCUAAUCA-------GUCGCAUUAUCAGCCCCAUUAUCAG ..(((((((((.((((.................(((((.....)))))---........--...((((((.....)))))).-------)))).)))....))))))....... ( -28.20) >DroSim_CAF1 1605 102 + 1 GUUGGGGUAAUCUGACACCUAUAUUGAUCUUCAGCCCAGUCAAUGGGC---AAUCACCC--AUCGAUUAGCAUUCCUAAUCA-------GCCGCAUUAUCAGCCCCAUUAUCAG ..(((((....((((..................(((((.....)))))---........--...((((((.....)))))).-------.........)))))))))....... ( -25.40) >DroEre_CAF1 1585 90 + 1 GUUUGUGCAAUCUGACACCUAUAUUGAUCUUCAGCCCAGUCAAUGGGC---AAUCAGCCGCAU--------------UAUCA-------GCCGUAUUAUCAACCGCAUUAUCAU (...((((...((((.........((.....))(((((.....)))))---..))))..))))--------------...).-------((.((.......)).))........ ( -17.60) >DroYak_CAF1 1588 104 + 1 GGUUGGGUAAUCUGACACCCAUAUUGAUCUUCAGCCCAGUCAAUGGGC---AAUCACCCCCAUCGAUUAGCACAGCUAAUCA-------GUCGCAUUAUCAGCGGCAUUAUCAU ((((((((........)))))............(((((.....)))))---....)))......(((((((...))))))).-------(((((.......)))))........ ( -31.80) >DroAna_CAF1 1984 110 + 1 GGCUGUGUAAUCAGAAUCCCAUUUUGAUCUUCAGCCCAAUCAAUGGGGUCCGGACA-CU--AUCGAUUAGUUACGGUUAUCAGCUAUCAGGCGUGUUAUCAAUAGCUGU-UCAA (((((....((((((((...))))))))...))))).((((.((((.((....)).-))--)).)))).(..((((((((..(((....))).((....))))))))))-.).. ( -24.90) >consensus GUUUGGGUAAUCUGACACCUAUAUUGAUCUUCAGCCCAGUCAAUGGGC___AAUCACCC__AUCGAUUAGCAUUCCUAAUCA_______GUCGCAUUAUCAGCCGCAUUAUCAG .....((((((.((..........((((.....(((((.....)))))....))))........((((((.....))))))..........)).)))))).............. (-11.51 = -13.32 + 1.81)
| Location | 14,325,074 – 14,325,176 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 76.97 |
| Mean single sequence MFE | -29.69 |
| Consensus MFE | -15.58 |
| Energy contribution | -17.30 |
| Covariance contribution | 1.72 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.52 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.507169 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
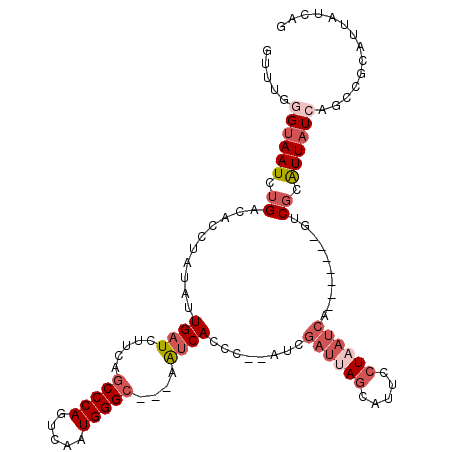
>2R_DroMel_CAF1 14325074 102 - 20766785 CUGAUAAUGCGGCUGAUAAUGCGAC-------UGAUUAGGAAUGCUAAUCGAU--GGGUGAUU---GCCCAUUGACUGGGCUGAAGAUCAAUAUAGGUGUCAGAUUACCGAAAC .((((....(((((.(.........-------.((((((.....))))))(((--((((....---)))))))...).)))))...)))).....((((......))))..... ( -28.40) >DroSec_CAF1 1605 102 - 1 CUGAUAAUGGGGCUGAUAAUGCGAC-------UGAUUAGGAAUGCUAAUCGUU--GGGUGAUU---GCCCAUUGACUGGGCUGAAGAUCAAUAUAGGUGUCAGAUUACCCCAAC .......(((((((((((((.((((-------.((((((.....)))))))))--).))((((---(((((.....)))))....))))........))))))....))))).. ( -33.10) >DroSim_CAF1 1605 102 - 1 CUGAUAAUGGGGCUGAUAAUGCGGC-------UGAUUAGGAAUGCUAAUCGAU--GGGUGAUU---GCCCAUUGACUGGGCUGAAGAUCAAUAUAGGUGUCAGAUUACCCCAAC .......(((((((((((.......-------(((((......(((..(((((--((((....---)))))))))...)))....))))).......))))))....))))).. ( -33.64) >DroEre_CAF1 1585 90 - 1 AUGAUAAUGCGGUUGAUAAUACGGC-------UGAUA--------------AUGCGGCUGAUU---GCCCAUUGACUGGGCUGAAGAUCAAUAUAGGUGUCAGAUUGCACAAAC .(((((.....((((((....((((-------((...--------------...))))))...---(((((.....))))).....)))))).....)))))............ ( -25.60) >DroYak_CAF1 1588 104 - 1 AUGAUAAUGCCGCUGAUAAUGCGAC-------UGAUUAGCUGUGCUAAUCGAUGGGGGUGAUU---GCCCAUUGACUGGGCUGAAGAUCAAUAUGGGUGUCAGAUUACCCAACC .((((...(((((.......))...-------.(((((((...)))))))((((((.(....)---.)))))).....))).....))))...((((((......))))))... ( -32.40) >DroAna_CAF1 1984 110 - 1 UUGA-ACAGCUAUUGAUAACACGCCUGAUAGCUGAUAACCGUAACUAAUCGAU--AG-UGUCCGGACCCCAUUGAUUGGGCUGAAGAUCAAAAUGGGAUUCUGAUUACACAGCC ....-.((((((((............))))))))...................--.(-(((.((((.((((((((((........)))))..)))))..))))...)))).... ( -25.00) >consensus CUGAUAAUGCGGCUGAUAAUGCGAC_______UGAUUAGGAAUGCUAAUCGAU__GGGUGAUU___GCCCAUUGACUGGGCUGAAGAUCAAUAUAGGUGUCAGAUUACCCAAAC ............((((((...............((((((.....))))))........(((((...(((((.....)))))....))))).......))))))........... (-15.58 = -17.30 + 1.72)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:17:42 2006