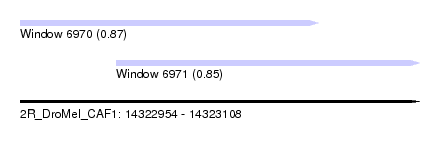
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,322,954 – 14,323,108 |
| Length | 154 |
| Max. P | 0.871325 |

| Location | 14,322,954 – 14,323,069 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.76 |
| Mean single sequence MFE | -20.19 |
| Consensus MFE | -3.10 |
| Energy contribution | -3.82 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.15 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.871325 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14322954 115 + 20766785 AAGACCGUUUCCAAAAACAA--UAAAAUAUCAUAGUUAA-AAAACUUCAGAGAAUGCAAAACGAAAGUGUUGGAAACAUUUCUAAGAUUAUGAUUG--AUCUUGAGCACUUUACUCAAAA ......((((((((......--.......((..((((..-..))))...)).........((....)).))))))))......(((((((....))--)))))(((.......))).... ( -19.80) >DroSec_CAF1 51968 115 + 1 AAGACCGUUUCCAAAAAUAA--AAAAAUGUCAUAGUCAA-AGAACUUCAGAGAAUGCAAAACGAUAGUGUUGGAAAUAUUUCUAAGAUUAUGAUUG--AUCUUGAGCACUGAACUCAAAA .(((..((((((((......--.....((.(((..((..-.........))..)))))...........))))))))...)))(((((((....))--)))))(((.......))).... ( -15.75) >DroSim_CAF1 53990 115 + 1 AAGACAGUUUCCAAAAAUAA--UAAAAUGUCAUAGUUAA-AAAACUUCAGAAAAUGCAACACGAAAGUGUUGUAAACAUUUCUAAGCUUAUGAUUG--AUCUUGAGCACUGAACUUAAAA ..((((..............--.....))))........-.....(((((....((((((((....))))))))...........(((((.((...--.)).))))).)))))....... ( -26.01) >DroEre_CAF1 52764 115 + 1 AACAACGUGUUUAAAAAUGAUAUAAAAUAUCAUAGGUAA-AUAAGAUCAGUAAAUGUAUAACUAAAACGGUGAAAGAA----UAAGAUUAUGCUUGUUACUUUGAGCACUGACAACCAAA .......((((((...(((((((...)))))))...)))-)))...(((((....((.........))(.(.((((((----((((......)))))).)))).).))))))........ ( -17.70) >DroYak_CAF1 53639 112 + 1 --------AUUUAGAAAUGAUCUAGAAUAUCAUAGUUCAAAAAGGAUCAGUAAAUGUAUAACUAGUUCGCUGCUUGAACUUCUAAGAUUAUGAUUCUUAAUUUGAGCACUGAACACAAAA --------(((((....((((((.((((......)))).....)))))).))))).........(((((.((((..((....(((((.......))))).))..)))).)))))...... ( -21.70) >consensus AAGACCGUUUCCAAAAAUAA__UAAAAUAUCAUAGUUAA_AAAACUUCAGAAAAUGCAAAACGAAAGUGUUGGAAAAAUUUCUAAGAUUAUGAUUG__AUCUUGAGCACUGAACUCAAAA ............................(((((((((..........((.....))(((.((....)).))).............))))))))).......................... ( -3.10 = -3.82 + 0.72)

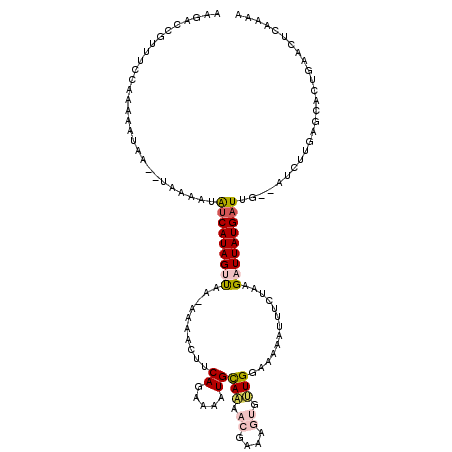
| Location | 14,322,991 – 14,323,108 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.67 |
| Mean single sequence MFE | -21.04 |
| Consensus MFE | -1.34 |
| Energy contribution | -2.70 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.06 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.851709 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14322991 117 + 20766785 AAAACUUCAGAGAAUGCAAAACGAAAGUGUUGGAAACAUUUCUAAGAUUAUGAUUG--AUCUUGAGCACUUUACUCAAAAUAGAUGAACGAAUAAU-UUUAAUUUACCUGACCUUCAAAA ......((((.(((......((....))..((....)).)))(((((((((..((.--((((((((.......))))....)))).))...)))))-))))......))))......... ( -18.90) >DroSec_CAF1 52005 117 + 1 AGAACUUCAGAGAAUGCAAAACGAUAGUGUUGGAAAUAUUUCUAAGAUUAUGAUUG--AUCUUGAGCACUGAACUCAAAAUAGAUGAACAAAUAAU-UUUAAUUUACCUGACCUUCAAAA .(((..((((....(.(((.((....)).))).)........(((((((((..((.--((((((((.......))))....)))).))...)))))-))))......))))..))).... ( -18.50) >DroSim_CAF1 54027 117 + 1 AAAACUUCAGAAAAUGCAACACGAAAGUGUUGUAAACAUUUCUAAGCUUAUGAUUG--AUCUUGAGCACUGAACUUAAAAUAAAUGAACGAGUAAU-UUUAAUUUACCUGACCUUCAAAA .....(((((....((((((((....))))))))...........(((((.((...--.)).))))).)))))...........((((((.((((.-......)))).))...))))... ( -25.70) >DroEre_CAF1 52803 115 + 1 AUAAGAUCAGUAAAUGUAUAACUAAAACGGUGAAAGAA----UAAGAUUAUGCUUGUUACUUUGAGCACUGACAACCAAAUAAAAGAUGGAAAUAU-UUGAAUUUUUAUGACCUUCAAAA ((((((....(((((((..........(((((((((((----((((......)))))).))))...)))))....(((.........)))..))))-)))...))))))........... ( -16.60) >DroYak_CAF1 53671 120 + 1 AAAGGAUCAGUAAAUGUAUAACUAGUUCGCUGCUUGAACUUCUAAGAUUAUGAUUCUUAAUUUGAGCACUGAACACAAAAUAAACCAACCACAUAUAUUAUAUUUAUCUGACCUUCAGAA .((((.((((((((((((......(((((.((((..((....(((((.......))))).))..)))).))))).....(((...........)))..)))))))).))))))))..... ( -25.50) >consensus AAAACUUCAGAAAAUGCAAAACGAAAGUGUUGGAAAAAUUUCUAAGAUUAUGAUUG__AUCUUGAGCACUGAACUCAAAAUAAAUGAACGAAUAAU_UUUAAUUUACCUGACCUUCAAAA ......((((......(((.((....)).))).............................(((((.......))))).............................))))......... ( -1.34 = -2.70 + 1.36)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:17:40 2006