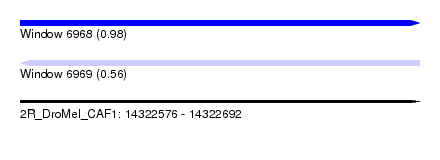
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,322,576 – 14,322,692 |
| Length | 116 |
| Max. P | 0.982164 |

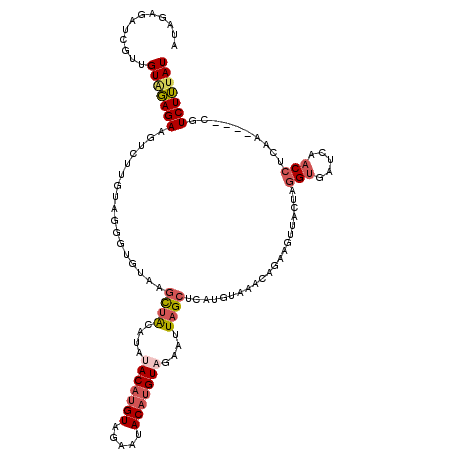
| Location | 14,322,576 – 14,322,692 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.32 |
| Mean single sequence MFE | -24.02 |
| Consensus MFE | -12.18 |
| Energy contribution | -14.40 |
| Covariance contribution | 2.22 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.51 |
| SVM decision value | 1.91 |
| SVM RNA-class probability | 0.982164 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14322576 116 + 20766785 AUAAAGACG----UUGAGGUUGAUCACCUGGUAACUUCUGUUUACAUGAGCUAAUUCUACAUGUAUUCUACAUGUAUAUGCAGCUUACACUGUACAAGAUUUCUCCACAACGAUCUCUAC ....(((((----(((((((.....))))((.....(((...((((((((((.....((((((((...)))))))).....))))))...))))..))).....)).))))).))).... ( -28.70) >DroSec_CAF1 51638 105 + 1 AUAAAGACG----UUGAGGUUGAUCACCUAGUAACUUCUGUUUACAUGGGCUAAUUCUAC-----------AUGUAUAUGUAGCUUACACCAUACAUGACAUCUCUACAACGAUCUCAAA .........----(((((((((....(((((((((....).)))).)))).........(-----------((((((.((((...))))..)))))))............))))))))). ( -23.10) >DroSim_CAF1 53645 116 + 1 AUAAAGACG----UUGAGGUUGAUCACCUAGUAACUUCUGUUUACAUGAGCUAAUUCUACAUGUAUUCUACAUGUAUAUGUAGCUUACACCAUACAAGACAUCUCUACAACGAUCUCAAA .........----(((((((((........(((((....).)))).(((((((....((((((((...))))))))....))))))).......................))))))))). ( -26.70) >DroEre_CAF1 52373 100 + 1 AUAAAGACAUUGGUUAAGGUUGAUCGCUCA-------------------GCGAAUUCUACAUGUAAUCUACAUGUCAAUGUAAUCUACACCCUACAAU-UUUCUCUACGACGAUCUCUGU ......((((((......(((((....)))-------------------)).......(((((((...))))))))))))).................-..................... ( -14.40) >DroYak_CAF1 53209 120 + 1 AUGUAGACAUUGGUUGUGGUUGAUCACCUAGCAACUUAUACUUACAUAAGCUAAUUCUACAUGUAAUCUACAUGUGUAUGUAGCUUACACUCUACAAGACUUCUAUACGGCAAACUCUAU .((((((.((.(((((((((.....)))..)))))).)).......(((((((....((((((((...))))))))....)))))))...))))))........................ ( -27.20) >consensus AUAAAGACG____UUGAGGUUGAUCACCUAGUAACUUCUGUUUACAUGAGCUAAUUCUACAUGUAAUCUACAUGUAUAUGUAGCUUACACCAUACAAGACUUCUCUACAACGAUCUCUAA ...............(((((((........((((.......)))).(((((((....(((((((.....)))))))....))))))).......................)))))))... (-12.18 = -14.40 + 2.22)

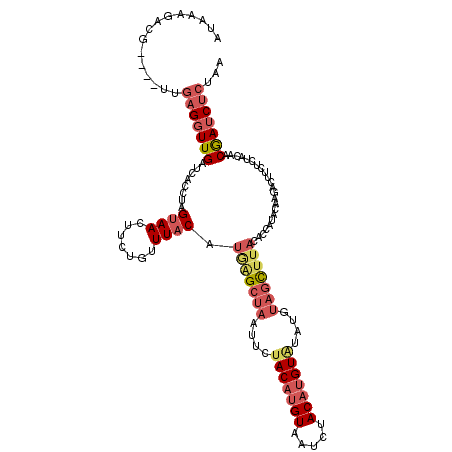
| Location | 14,322,576 – 14,322,692 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.32 |
| Mean single sequence MFE | -27.08 |
| Consensus MFE | -11.30 |
| Energy contribution | -12.44 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.42 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.558200 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14322576 116 - 20766785 GUAGAGAUCGUUGUGGAGAAAUCUUGUACAGUGUAAGCUGCAUAUACAUGUAGAAUACAUGUAGAAUUAGCUCAUGUAAACAGAAGUUACCAGGUGAUCAACCUCAA----CGUCUUUAU (((((((.((((((((.((..(((..((((.((..(((((....((((((((...))))))))....)))))))))))...)))..)).)))(((.....))).)))----))))))))) ( -33.70) >DroSec_CAF1 51638 105 - 1 UUUGAGAUCGUUGUAGAGAUGUCAUGUAUGGUGUAAGCUACAUAUACAU-----------GUAGAAUUAGCCCAUGUAAACAGAAGUUACUAGGUGAUCAACCUCAA----CGUCUUUAU ...((((.((((((((.(((.((.(((((((.((...((((((....))-----------)))).....))))))....))))).))).)))(((.....))).)))----))))))... ( -23.70) >DroSim_CAF1 53645 116 - 1 UUUGAGAUCGUUGUAGAGAUGUCUUGUAUGGUGUAAGCUACAUAUACAUGUAGAAUACAUGUAGAAUUAGCUCAUGUAAACAGAAGUUACUAGGUGAUCAACCUCAA----CGUCUUUAU ...((((.((((((((.(((.(((.((...(..(.(((((....((((((((...))))))))....))))).)..)..))))).))).)))(((.....))).)))----))))))... ( -29.40) >DroEre_CAF1 52373 100 - 1 ACAGAGAUCGUCGUAGAGAAA-AUUGUAGGGUGUAGAUUACAUUGACAUGUAGAUUACAUGUAGAAUUCGC-------------------UGAGCGAUCAACCUUAACCAAUGUCUUUAU ............(((((((..-((((((((((((((.((((((....)))))).)))).....((..((((-------------------...)))))).))))))..)))).))))))) ( -20.80) >DroYak_CAF1 53209 120 - 1 AUAGAGUUUGCCGUAUAGAAGUCUUGUAGAGUGUAAGCUACAUACACAUGUAGAUUACAUGUAGAAUUAGCUUAUGUAAGUAUAAGUUGCUAGGUGAUCAACCACAACCAAUGUCUACAU ..(((.(((........))).)))(((((((..(((((((.....(((((((...))))))).....)))))))..)........((((...(((.....))).)))).....)))))). ( -27.80) >consensus AUAGAGAUCGUUGUAGAGAAGUCUUGUAGGGUGUAAGCUACAUAUACAUGUAGAAUACAUGUAGAAUUAGCUCAUGUAAACAGAAGUUACUAGGUGAUCAACCUCAA____CGUCUUUAU ............(((((((.................((((....(((((((.....)))))))....)))).....................(((.....)))..........))))))) (-11.30 = -12.44 + 1.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:17:38 2006