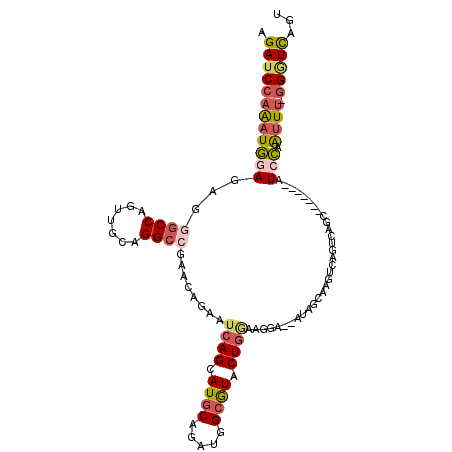
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 2,511,015 – 2,511,116 |
| Length | 101 |
| Max. P | 0.990060 |

| Location | 2,511,015 – 2,511,116 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 76.42 |
| Mean single sequence MFE | -29.89 |
| Consensus MFE | -17.33 |
| Energy contribution | -17.13 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.58 |
| SVM decision value | 2.19 |
| SVM RNA-class probability | 0.990060 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
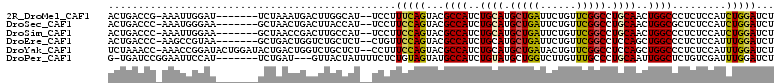
>2R_DroMel_CAF1 2511015 101 + 20766785 ACUGACCG-AAAUUGGAU-------UCUAAAUGACUUGGCAU--UCCUUUCAGUACGCCAUCUGCAUGCUGAUUCUGUUCGGCCUGCAACUGGCCCUCUCCAUCUGGAUCU (((((...-.....(((.-------.((((.....))))...--)))..)))))..((((..((((.(((((......))))).))))..))))....(((....)))... ( -27.00) >DroSec_CAF1 20627 101 + 1 ACUGACCC-AAAUGGGAA-------GCUAACUGACUUACCAU--UCCUUCCAGUACGCCAUCUGCAUGCUGAUUCUGUUCGGCCUGCAACUGGCGCUCUCCAUCUGGAUCU ...((.((-(.((((..(-------((..((((.........--......))))..((((..((((.(((((......))))).))))..)))))))..)))).))).)). ( -37.66) >DroSim_CAF1 20753 101 + 1 ACUGACCC-AAAUUGGAA-------GCUAACCGACUUGCCAU--UCCUUCCAGUACGCCAUCUGCAUGCUGAUUCUGUUCGGCCUGCAACUGGCCCUCUCCAUCUGGAUCU ...((.((-(.(((((((-------(................--..))))))))..((((..((((.(((((......))))).))))..))))..........))).)). ( -29.67) >DroEre_CAF1 20849 101 + 1 ACUGACCC-AAGCCGUAA-------GCUGACUGGUCUGCUCU--CUGUUCCAGUACGCCAUCUGCAUGCUGAUUCUGUUCGGCCUCCAGCUGGCCCUCUCCAUUUGGAUCU ...((.((-(((......-------....(((((...((...--..)).)))))..((((.(((...(((((......)))))...))).))))........))))).)). ( -28.30) >DroYak_CAF1 20067 108 + 1 UCUAAACC-AAACCGGAUACUGGAUACUGACUGGUCUGCUCU--CCUUUCCAGUACGCCAUCUGCAUGCUGAUACUGUUCGGCCUCCAGCUGGCCCUCUCCAUUUGGAUCU ......((-(((..((((((((((....(((......).)).--....))))))).((((.(((...(((((......)))))...))).))))....))).))))).... ( -32.50) >DroPer_CAF1 22634 100 + 1 G-UGAUCCGGAAUUCCAU-------UCUGAU---GUUACUAUUUUCUCUGUAGUAUGCCAUCUGUAUGCUGGUCUUGUUUGCCCUGCAAUUGGCUCUGUCGAUUUGGAUCU .-.(((((((....))..-------..((((---..((((((.......)))))).((((..((((....(((.......))).))))..))))...))))....))))). ( -24.20) >consensus ACUGACCC_AAAUUGGAA_______GCUAACUGACUUGCCAU__UCCUUCCAGUACGCCAUCUGCAUGCUGAUUCUGUUCGGCCUGCAACUGGCCCUCUCCAUCUGGAUCU ................................................(((((...((((..((((.(((((......))))).))))..)))).........)))))... (-17.33 = -17.13 + -0.19)
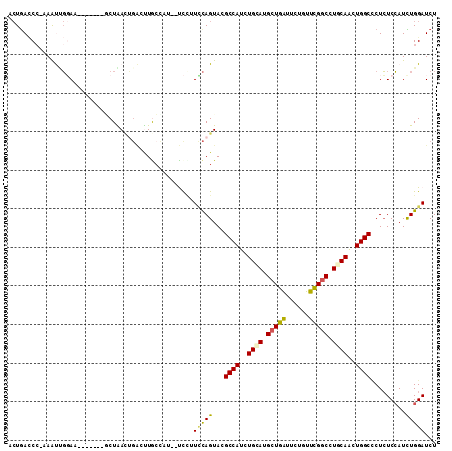
| Location | 2,511,015 – 2,511,116 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 76.42 |
| Mean single sequence MFE | -31.70 |
| Consensus MFE | -16.94 |
| Energy contribution | -17.70 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.881566 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
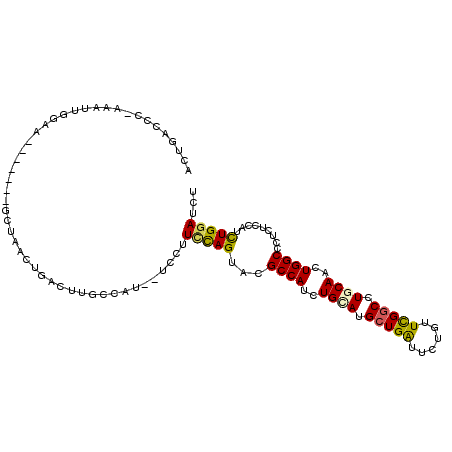
>2R_DroMel_CAF1 2511015 101 - 20766785 AGAUCCAGAUGGAGAGGGCCAGUUGCAGGCCGAACAGAAUCAGCAUGCAGAUGGCGUACUGAAAGGA--AUGCCAAGUCAUUUAGA-------AUCCAAUUU-CGGUCAGU .((((.(((((((...(((...(((((.((.((......)).)).))))).((((((.((....)).--)))))).))).......-------.))).))))-.))))... ( -28.20) >DroSec_CAF1 20627 101 - 1 AGAUCCAGAUGGAGAGCGCCAGUUGCAGGCCGAACAGAAUCAGCAUGCAGAUGGCGUACUGGAAGGA--AUGGUAAGUCAGUUAGC-------UUCCCAUUU-GGGUCAGU .(((((((((((..(((((((.(((((.((.((......)).)).))))).))))..(((((.....--........)))))..))-------)..))))))-)))))... ( -42.52) >DroSim_CAF1 20753 101 - 1 AGAUCCAGAUGGAGAGGGCCAGUUGCAGGCCGAACAGAAUCAGCAUGCAGAUGGCGUACUGGAAGGA--AUGGCAAGUCGGUUAGC-------UUCCAAUUU-GGGUCAGU .(((((((((..((.(.((((.(((((.((.((......)).)).))))).)))).).))(((((..--.((((......)))).)-------)))).))))-)))))... ( -36.70) >DroEre_CAF1 20849 101 - 1 AGAUCCAAAUGGAGAGGGCCAGCUGGAGGCCGAACAGAAUCAGCAUGCAGAUGGCGUACUGGAACAG--AGAGCAGACCAGUCAGC-------UUACGGCUU-GGGUCAGU .(((((...(((......))).....((((((.......((((.((((.....)))).)))).....--.((((.((....)).))-------)).))))))-)))))... ( -30.60) >DroYak_CAF1 20067 108 - 1 AGAUCCAAAUGGAGAGGGCCAGCUGGAGGCCGAACAGUAUCAGCAUGCAGAUGGCGUACUGGAAAGG--AGAGCAGACCAGUCAGUAUCCAGUAUCCGGUUU-GGUUUAGA .((.(((((((((..((((((.(((.(.((.((......)).)).).))).))))(((((((...((--........))..)))))))))....))).))))-)).))... ( -31.40) >DroPer_CAF1 22634 100 - 1 AGAUCCAAAUCGACAGAGCCAAUUGCAGGGCAAACAAGACCAGCAUACAGAUGGCAUACUACAGAGAAAAUAGUAAC---AUCAGA-------AUGGAAUUCCGGAUCA-C .(((((.........((((((..(((.((..........)).)))......)))).(((((.........)))))..---.))...-------..((....))))))).-. ( -20.80) >consensus AGAUCCAAAUGGAGAGGGCCAGUUGCAGGCCGAACAGAAUCAGCAUGCAGAUGGCGUACUGGAAGGA__AUAGCAAGUCAGUCAGC_______AUCCAAUUU_GGGUCAGU .((((((((((((...((((.......))))........((((.((((.....)))).))))................................))).)))).)))))... (-16.94 = -17.70 + 0.76)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:38:31 2006