| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,299,242 – 14,299,347 |
| Length | 105 |
| Max. P | 0.960075 |

| Location | 14,299,242 – 14,299,347 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.81 |
| Mean single sequence MFE | -34.68 |
| Consensus MFE | -20.88 |
| Energy contribution | -19.85 |
| Covariance contribution | -1.03 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.87 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.960075 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
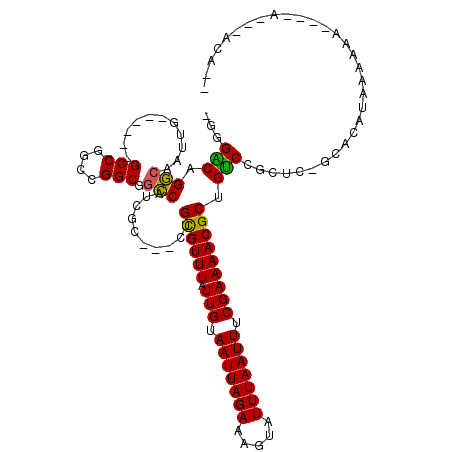
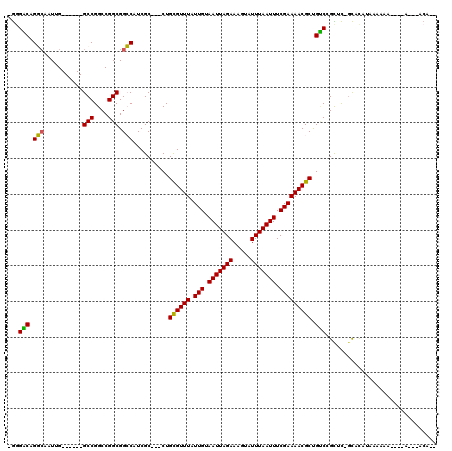
>2R_DroMel_CAF1 14299242 105 - 20766785 -GCGACAGGCAAUUG------GCCGGCCGGCGGCCAUCGC---CUGCGUUUAUUGUAAUUAGAAAGUAUUUAAUUUCGAAAACGCUGUCCGCUC-GCACAUAAAAAA----AAAAACAAA -(((((((((...((------((((.....))))))..))---))((((((.(((.(((((((.....))))))).))))))))).....).))-))..........----......... ( -33.50) >DroPse_CAF1 31207 108 - 1 -GGGGCAGCCAAUUG------GCCGGCCGGCGGGCAUCGCCGUCUGCGUUUAUUGUAAUUAGAAAGUAUUUAAUUUCGAAAACGCCGCCAGUUC-GCAUAUAAAAAUGUGUA--UACA-- -..(((.(((....)------))..)))(((((((...)))....((((((.(((.(((((((.....))))))).))))))))))))).((..-((((((....)))))).--.)).-- ( -35.80) >DroEre_CAF1 29080 100 - 1 -GGGACAGGCAAUUG------GCCGGCCGGCGGCCGUCGC---CUGUGUUUAUUGUAAUUAGAAAGUAUUUAAUUUCGAAAACGCUGUCCGCUC-GCACAUAAAAAA---------CAAA -.((((((((...((------((((.....))))))..))---))((((((.(((.(((((((.....))))))).))))))))).))))....-............---------.... ( -31.90) >DroYak_CAF1 29268 115 - 1 -GGGACAGGCAAUUGGCAAUGGCCGGCCGGCGGCCAUCGC---CUGCGUUUAUUGUAAUUAGAAAGUAUUUAAUUUCGAAAACGCUGUCUGCUC-GCACAUAAAAAACUAAAACAACAAU -(((.((((((...(((.(((((((.....))))))).))---).((((((.(((.(((((((.....))))))).))))))))))))))))))-......................... ( -39.70) >DroAna_CAF1 35173 93 - 1 CAGGUCUGGACAUUG------GCCGGCCGGCGGCCAUCGC---CUGCGUUUAUUGUAAUUAGAAAGUAUUUAAUUUCGAAAACGCUGACCGUUUUUUACAUA------------------ ..((((.((....((------((((.....))))))...)---).((((((.(((.(((((((.....))))))).))))))))).))))............------------------ ( -30.10) >DroPer_CAF1 31254 108 - 1 -GGGGCAGCCAAUUG------GCCGGCCGGCGGGCAUCGCCGCCUGCGUUUAUUGUAAUUAGAAAGUAUUUAAUUUCGAAAACGCCGCCAGUUC-GCAUAUAAAAAUGUGUA--UACA-- -..(((.(((....)------))..)))(((((((......))).((((((.(((.(((((((.....))))))).))))))))))))).((..-((((((....)))))).--.)).-- ( -37.10) >consensus _GGGACAGGCAAUUG______GCCGGCCGGCGGCCAUCGC___CUGCGUUUAUUGUAAUUAGAAAGUAUUUAAUUUCGAAAACGCUGUCCGCUC_GCACAUAAAAAA____A___ACA__ ...(((.(((...........(((....))).)))..........((((((.(((.(((((((.....))))))).))))))))).)))............................... (-20.88 = -19.85 + -1.03)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:17:33 2006