| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,295,452 – 14,295,551 |
| Length | 99 |
| Max. P | 0.953296 |

| Location | 14,295,452 – 14,295,551 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 76.51 |
| Mean single sequence MFE | -17.64 |
| Consensus MFE | -9.91 |
| Energy contribution | -9.83 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.953296 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14295452 99 - 20766785 UUCUCCGCAUUUCUCUUUCCUCAACUGGAUUGUUGUUUUUUUUUUGGUUCACUCUGCCAACUGCAAUUAAAAUUUUGUAAUACGGUGACCAAAAA------AGCA---- ......((......(..(((......)))..).......(((((((((.((((.((.....(((((........))))).)).))))))))))))------))).---- ( -19.10) >DroSec_CAF1 24638 88 - 1 UUCUCCGCAUUUCUCUUUCCUCAA------------UUUU-UUUUGGUUCACUCUGCCAACUACAAUUAAAAUUUUGUAAUACGGUGACCAAAAA--AA--AAGA---- ........................------------((((-(((((((.((((.((.....(((((........))))).)).))))))))))))--))--)...---- ( -15.40) >DroSim_CAF1 24713 90 - 1 UUCUCCGCAUUUCUCUUUCCUCAA------------UUUUUUUUUGGUUCACUCUGCCAACUGCAAUUAAAAUUUUGUAAUACGGUGACCAAAAA-AAA--AAGA---- ........................------------((((((((((((.((((.((.....(((((........))))).)).))))))))))))-)))--)...---- ( -16.30) >DroEre_CAF1 25244 90 - 1 ---------UUUCUCUUUCGGCAGCUGCGC---UUU----UUUUUGGUUCACUCUGCCAACUGCAAUUAAAAUUUUGUAAUACGGUGACCAAAAA-AGA--AAGAAAAG ---------....(((((((((......))---).(----((((((((.((((.((.....(((((........))))).)).))))))))))))-)))--)))).... ( -21.40) >DroYak_CAF1 25360 106 - 1 UUCUCCGCAUUUCACUUUCAUCAACUGAGU---UUUUUUUUUUUUGGUUCACUCUGCCAGCUGCAAUUAAAAUUUUGUAAUACGGUGACCAAAAACAGAAAAAAAACAG ........................(((..(---(((((((((((((((.((((.((.....(((((........))))).)).)))))))))))).))))))))..))) ( -19.40) >DroAna_CAF1 31212 95 - 1 UUCUCCGCUCUUUACUUUCCUCAACUGCAC--------UUUUUUGGCUUUACUCCGCCGACUGCAAUUAAAAUUUUGUAAUACGGUGACCAAAGA------AACAAAAA ......((..................))..--------.((((((.((((....(((((..(((((........)))))...)))))...)))).------..)))))) ( -14.27) >consensus UUCUCCGCAUUUCUCUUUCCUCAACUG_________UUUUUUUUUGGUUCACUCUGCCAACUGCAAUUAAAAUUUUGUAAUACGGUGACCAAAAA___A__AAGA____ .........................................(((((((.((((........(((((........)))))....)))))))))))............... ( -9.91 = -9.83 + -0.08)

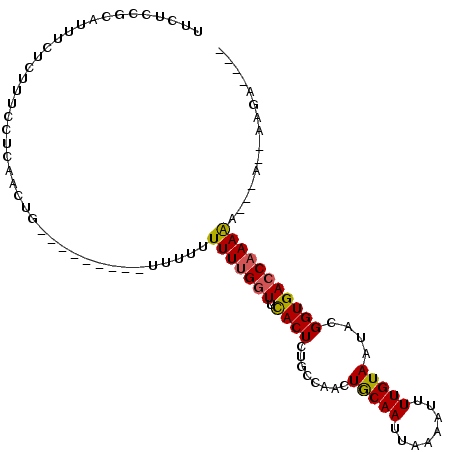
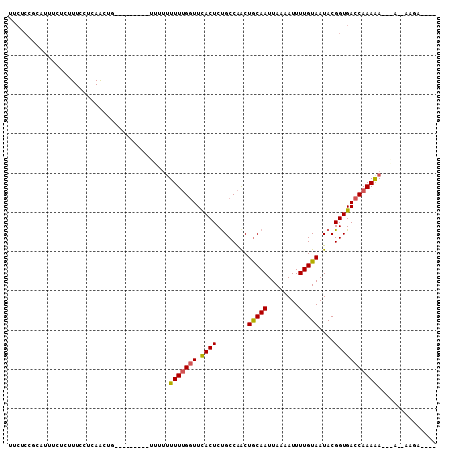
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:17:31 2006