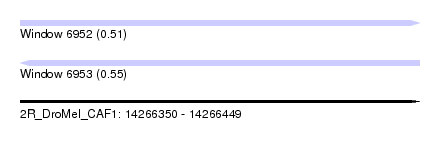
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,266,350 – 14,266,449 |
| Length | 99 |
| Max. P | 0.553349 |

| Location | 14,266,350 – 14,266,449 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 83.33 |
| Mean single sequence MFE | -36.35 |
| Consensus MFE | -22.48 |
| Energy contribution | -22.53 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.62 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.512759 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14266350 99 + 20766785 -AAAGGAGCCAAGGACUC---UGUUUAACAUGGUCGUAGAGCUGGCUGAAGU--------GGGGAGCUG-GGAGCUGCUCAUUUGCAGCUGCUGGCAAUUCAAUUUGCCAUC -.....((((.(.((((.---((.....)).)))).).).)))(((.....(--------(((..((..-(.((((((......)))))).)..))..))))....)))... ( -37.10) >DroSec_CAF1 3626 98 + 1 -AAAGGAGCCAAGGACUC---UGUUUAACAUGGUCGUGGAGCUGGCUGGAGU--------GGGUAG-UG-GGAGCUGCUCAUUUGCAGCUGCUGGCAAUUCAAUUUGCCAUC -.......(((..((((.---((.....)).)))).)))(((.(((((((((--------((((((-(.-...))))))))))).))))))))(((((......)))))... ( -39.00) >DroSim_CAF1 3731 98 + 1 -AAAGGAGCCAAGGACUC---UGUUUAACAUGGUCGUGGAGCUGGCUGGAGU--------GGGUAG-UG-GGAGCUGCUCAUUUGCAGCUGCUGGCAAUUCAAUUUGCCAUC -.......(((..((((.---((.....)).)))).)))(((.(((((((((--------((((((-(.-...))))))))))).))))))))(((((......)))))... ( -39.00) >DroEre_CAF1 3574 104 + 1 -AACGGAGCCAAGGACUC---UGUUUAACAUGGUCGUCG-GCGAGCUGGAGUAGG-GAAAGGGAAG-UG-GGAGCUGCUCAUUUGCAGCUGCUGGCAAUUCAAUUUGCCAUC -..(((.(((.(.((((.---((.....)).)))).).)-))...))).....((-.(((..((((-(.-((((((((......)))))).)).))..)))..))).))... ( -36.30) >DroYak_CAF1 3650 103 + 1 -AAAGGAGCCAAGGACUC---UGUUUAACAUGGUCGUC----CAGCUGGAGUGGGAGUGAGGGGAG-UGGGGAGCUGCUCAUUUGCAGCUGCUGGCAAUUCAAUUUGCCAUC -..((.(((....((((.---((.....)).)))).((----(.((....)).)))(..((..(((-..(....)..))).))..).))).))(((((......)))))... ( -32.40) >DroAna_CAF1 4097 94 + 1 AAAAGGAGCCAAGGACCUGAAGGCUUGACAUGGCCGUGAAG--GGCUGA--------------CAG-GA-GGAGCUGCUCAUUUGCAGCUGUUGGCCAUUCAAUUUGCCAUC .....(((((...........)))))...(((((..((((.--(((..(--------------((.-..-...(((((......))))))))..))).))))....))))). ( -34.30) >consensus _AAAGGAGCCAAGGACUC___UGUUUAACAUGGUCGUGGAGCUGGCUGGAGU________GGGGAG_UG_GGAGCUGCUCAUUUGCAGCUGCUGGCAAUUCAAUUUGCCAUC .......((((.((((......))))....))))....................................(.((((((......)))))).)((((((......)))))).. (-22.48 = -22.53 + 0.06)

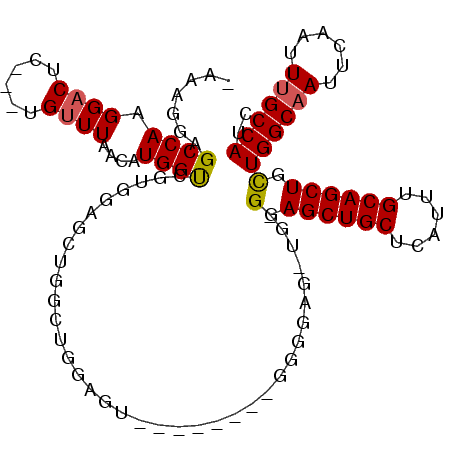
| Location | 14,266,350 – 14,266,449 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 83.33 |
| Mean single sequence MFE | -26.62 |
| Consensus MFE | -18.99 |
| Energy contribution | -19.18 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.553349 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14266350 99 - 20766785 GAUGGCAAAUUGAAUUGCCAGCAGCUGCAAAUGAGCAGCUCC-CAGCUCCCC--------ACUUCAGCCAGCUCUACGACCAUGUUAAACA---GAGUCCUUGGCUCCUUU- ..((((((......))))))((((((((......))))))..-..)).....--------.....((((((((((..(((...)))....)---))))...))))).....- ( -26.30) >DroSec_CAF1 3626 98 - 1 GAUGGCAAAUUGAAUUGCCAGCAGCUGCAAAUGAGCAGCUCC-CA-CUACCC--------ACUCCAGCCAGCUCCACGACCAUGUUAAACA---GAGUCCUUGGCUCCUUU- ..((((((......))))))..((((((......))))))..-..-......--------.....((((((......(((..((.....))---..))).)))))).....- ( -24.70) >DroSim_CAF1 3731 98 - 1 GAUGGCAAAUUGAAUUGCCAGCAGCUGCAAAUGAGCAGCUCC-CA-CUACCC--------ACUCCAGCCAGCUCCACGACCAUGUUAAACA---GAGUCCUUGGCUCCUUU- ..((((((......))))))..((((((......))))))..-..-......--------.....((((((......(((..((.....))---..))).)))))).....- ( -24.70) >DroEre_CAF1 3574 104 - 1 GAUGGCAAAUUGAAUUGCCAGCAGCUGCAAAUGAGCAGCUCC-CA-CUUCCCUUUC-CCUACUCCAGCUCGC-CGACGACCAUGUUAAACA---GAGUCCUUGGCUCCGUU- ..((((((......))))))..((((((......))))))..-..-..........-.............((-(((.(((..((.....))---..))).)))))......- ( -28.50) >DroYak_CAF1 3650 103 - 1 GAUGGCAAAUUGAAUUGCCAGCAGCUGCAAAUGAGCAGCUCCCCA-CUCCCCUCACUCCCACUCCAGCUG----GACGACCAUGUUAAACA---GAGUCCUUGGCUCCUUU- ..((((((......))))))..((((((......)))))).....-...................(((((----(((.....((.....))---..))))..)))).....- ( -24.70) >DroAna_CAF1 4097 94 - 1 GAUGGCAAAUUGAAUGGCCAACAGCUGCAAAUGAGCAGCUCC-UC-CUG--------------UCAGCC--CUUCACGGCCAUGUCAAGCCUUCAGGUCCUUGGCUCCUUUU .(((((....((((.(((..((((((((......)))))...-..-.))--------------)..)))--.))))..)))))((((((((....))..))))))....... ( -30.80) >consensus GAUGGCAAAUUGAAUUGCCAGCAGCUGCAAAUGAGCAGCUCC_CA_CUACCC________ACUCCAGCCAGCUCCACGACCAUGUUAAACA___GAGUCCUUGGCUCCUUU_ ..((((((......))))))..((((((......))))))......................................................((((.....))))..... (-18.99 = -19.18 + 0.20)
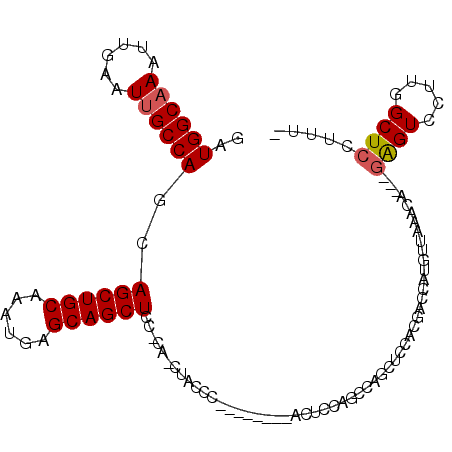
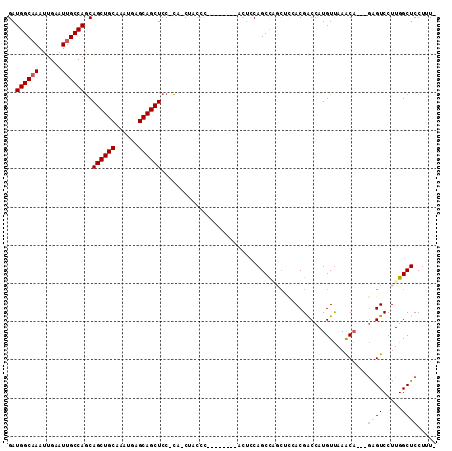
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:17:22 2006