| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,244,764 – 14,244,884 |
| Length | 120 |
| Max. P | 0.601365 |

| Location | 14,244,764 – 14,244,884 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.78 |
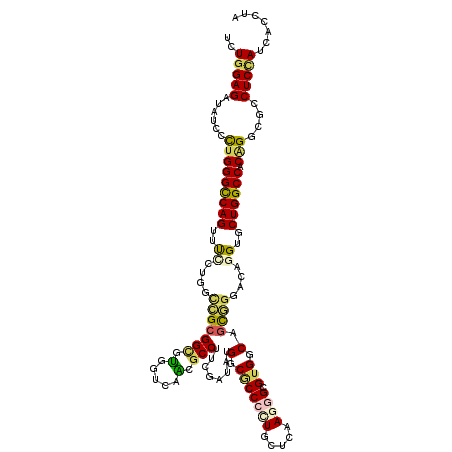
| Mean single sequence MFE | -55.77 |
| Consensus MFE | -32.27 |
| Energy contribution | -32.00 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.601365 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
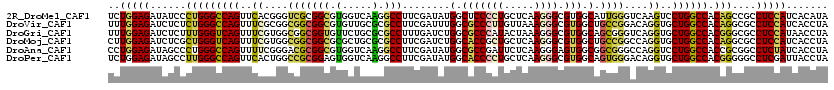
>2R_DroMel_CAF1 14244764 120 - 20766785 UCUGGAGAUAUCCCUGGGCCAGUUCACGGGUCGCGGCGUGGUCAAGGCCUUCGAUAUGGCUCCCCUGCUCAAGGGCGUGGCAUUGGGUCAAGUCCUGGCCACAGCCGCCUCCAUCACAUA ..(((((....(((((((....)))).)))..((((((((((((.((.(((.((((((.(.(((((.....)))).).).)))...)))))).))))))))).))))))))))....... ( -50.10) >DroVir_CAF1 1558 120 - 1 UUUGGAGAUCUCUCUGGGCCAGUUUCGCGGCGGCGGCGUGUUGCGCGCCUUCGAUUUGGCGCCCUUGUUAAAGGGCGUGGCUGCCGGACAGGUGCUGGCCACAGGCGCCUCCAUCACCUA ..(((((.....(((((((((((.((.(((((((((((((...)))))).........((((((((....)))))))).)))))))....)).))))))).))))...)))))....... ( -61.90) >DroGri_CAF1 15325 120 - 1 UUUGGAGAUCUCUUUGGGUCAGUUUCGUGGCGGCGGUGUUCUGCGCGCCUUUGAUCUGGCGCCCAUACUAAAGGGCGUGGCAGCGGGUCAGGUGCUGGCCACGGGCGCCUCCAUAACCUA ..(((((((((....))))).(((.((((((.((((....))))((((((..((((((((((((........)))))).....))))))))))))..)))))))))...))))....... ( -51.20) >DroMoj_CAF1 2278 120 - 1 CUUGGAGAUCUCGCUGGGUCAGUUUCGUGGCGGCGGCGCGCUGCGCGCCUUCGAUCUGGCACCGCUGCUCAAGGGCGUGGCUGCCGGCCAGGUGCUGGCCACAGGCGCCUCCAUCACCUA ..(((((....((((.(((((((..(.((((((.((((((...)))))).))....(((((((((.(((....))))))).))))))))).).)))))))...)))).)))))....... ( -60.50) >DroAna_CAF1 1546 120 - 1 CCUGGAGAUAGCCCUGGGCCAGUUUUCGGGACGCGGCGUGGUCAAGGCCUUCGAUAUGGCGCCGAUUCUCAAGGGAGUGGCGGCGGGCCAGGUCCUGGCCACCGCGGCCUCUAUCACCUA ...((.(((((....(((((.(((.....)))((((..((((((.(((((..(...((.((((.(((((....))))))))).))..).))))).)))))))))))))))))))).)).. ( -54.40) >DroPer_CAF1 1441 120 - 1 UCUGGAGAUAGCCUUGGGCCAGUUCACUGGCCGCGGAGUGGUCAAGGCCUUCGAUAUGGCACCCCUGCUCAAGGGCGUGGCAGUGGGACAGGUGCUGGCCACGGGGGCCUCGAUUACCUA (((((((.....))).((((((....)))))).))))((((((.((((((((....((.(((((((.....)))).))).))((((..(((...))).)))))))))))).))))))... ( -56.50) >consensus UCUGGAGAUAUCCCUGGGCCAGUUUCCUGGCCGCGGCGUGGUCAACGCCUUCGAUAUGGCGCCCCUGCUCAAGGGCGUGGCAGCGGGACAGGUGCUGGCCACAGGCGCCUCCAUCACCUA ..(((((......(((((((((..((....(((((((.(.....).)))........(.(((((((.....)))).))).).))))....))..)))))).)))....)))))....... (-32.27 = -32.00 + -0.27)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:17:17 2006