| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,244,578 – 14,244,684 |
| Length | 106 |
| Max. P | 0.628763 |

| Location | 14,244,578 – 14,244,684 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 73.86 |
| Mean single sequence MFE | -30.36 |
| Consensus MFE | -16.97 |
| Energy contribution | -17.78 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.628763 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
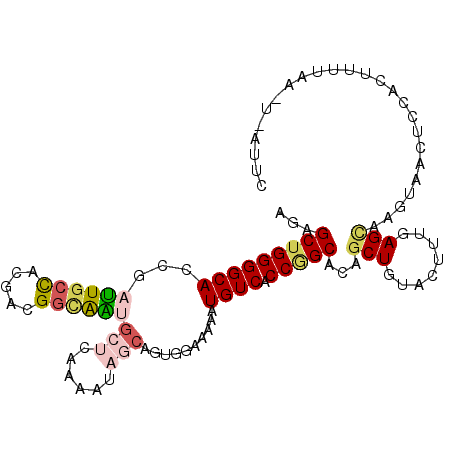
>2R_DroMel_CAF1 14244578 106 - 20766785 AGAGCUGGGGCACCGAUUGCCACGACGGCAAUGCUCAAAAUAGCAGUGGAAAAAUGUCACCGGCACAGCUGUACUUUGAGCAAGUACCUCCACUUUUAAAUCAUUC ((((.(((((.((..((((((.....))))))((((((((((((.(((...............))).)))))..)))))))..)).)).))))))).......... ( -30.26) >DroVir_CAF1 1372 106 - 1 AGAGCUGGGGCAGCGAUUGCUACGAUGGUGGCUCGGUAAAUGCCACUGGUAAAAUGUCACCCGCGCAGCUGUACUUUGAGUGAGUAAACAUAUCUGGAGGGUAUGU ..((((((((..(((.(((((.....((((((.........)))))))))))..)))..)))...))))).(((((.....))))).(((((((.....))))))) ( -30.70) >DroSec_CAF1 1356 82 - 1 AGAGCUGGGGCACCGAUUGCCACGACGGCAAUGCUCAAAAUAGCAGUGGAAAAAUGUCACCGGCACAGCUGUACUUUGAG------------------------UC ...((((.((((.....))))....))))...((((((((((((.(((...............))).)))))..))))))------------------------). ( -23.56) >DroSim_CAF1 1430 106 - 1 AGAGCUGGGGCACCGAUUGCCACGACGGCAAUGCUCAAAAUAGCAGUGGAAAAAUGUCACCGGCACAGCUGUACUUUGAGCAAGUAUCUCCACUCUUAAUUCAUUC ((((.(((((.....((((((.....))))))((((((((((((.(((...............))).)))))..)))))))......))))))))).......... ( -32.06) >DroYak_CAF1 1473 106 - 1 AGAGCUGGGGCACCGAUUGCCACGAUGGCAAUGCUCAAAAUAGCAGUGGAAAAAUGUCACCGGCACAGCUGUACUUUGAGCAAGUACCUCCACUUUUAAUUAAUUC ((((.(((((.((..((((((.....))))))((((((((((((.(((...............))).)))))..)))))))..)).)).))))))).......... ( -30.56) >DroMoj_CAF1 2108 90 - 1 CGAGCUGGGGCAGCGACUGCCACGAUGUGAGCGCGGCGAAUGCCAGUGGCAAAAUGUCACCAGCUCAGCUGUAUUUUGAGUGAGUGAAGA---------------- .(((((((((((.....((((((...((....))(((....))).))))))...)))).)))))))..((.(((((.....))))).)).---------------- ( -35.00) >consensus AGAGCUGGGGCACCGAUUGCCACGACGGCAAUGCUCAAAAUAGCAGUGGAAAAAUGUCACCGGCACAGCUGUACUUUGAGCAAGUAACUCCACUUUUAA_U_AUUC ...(((((((((...((((((.....))))))(((......)))..........)))).)))))...(((........)))......................... (-16.97 = -17.78 + 0.81)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:17:16 2006