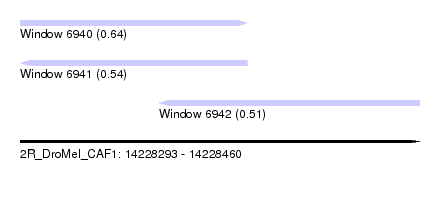
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,228,293 – 14,228,460 |
| Length | 167 |
| Max. P | 0.644171 |

| Location | 14,228,293 – 14,228,388 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 78.92 |
| Mean single sequence MFE | -27.67 |
| Consensus MFE | -13.18 |
| Energy contribution | -13.52 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.644171 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14228293 95 + 20766785 CCAU-GGU---UUUUAACCACGACUUUAAGUGCGACGAUAAGGUUGUCAAGUGCGGACAGUUUUAUCAGGCCGCACAG-CAAAUGCAAUUUAAAUGCAUU ...(-(((---.....)))).........(((((.((((((..(((((.......)))))..))))).)..)))))..-..((((((.......)))))) ( -26.00) >DroSim_CAF1 9246 95 + 1 CCAU-GGU---UUUUAACCACGAGUUUAAGUGCGGCGAUAAGGUUGUCAAGUGCGGACAGUUUUAUCAGGCCGUACGG-CAAAUGCAAUUUAAAUGCAUU ((.(-(((---.....)))).........((((((((((((..(((((.......)))))..)))))..)))))))))-..((((((.......)))))) ( -31.00) >DroEre_CAF1 8011 95 + 1 GCAU-GGU---UUUUAACCACGAGCUUAAGUGCGGCGAUAAGGUUGUCAAGUGCGGACAGUUUUAUCAGCCCGCACAG-CAAAUGCAAUUUAAAUGCAUU ((.(-(((---.....)))).........((((((((((((..(((((.......)))))..))))).).)))))).)-).((((((.......)))))) ( -31.90) >DroYak_CAF1 7705 87 + 1 GCAU-GGU---UUUUUACCACGGGUUUAAGUGCGCCGAUAAGGUUGUCAAGUGCGGACAGUUUUAUCAGC---------CAAAUGCAAUUUAAAUGCAUU ...(-(((---.....)))).(.((((((((((((.(((((..(((((.......)))))..))))).))---------.....)).)))))))).)... ( -23.80) >DroAna_CAF1 13533 90 + 1 GAAU-GGU---UUUUU-CC---ACUUUGGGCGUUGCGAUAAGGUUGUCAAGUGCCGACAGGUUUAUCUU-CCGCCAAA-GAAACGCAAUUUAAAUGCAUU ...(-((.---.....-))---)((((((.((..(.((((((.(((((.......))))).))))))).-.)))))))-)....(((.......)))... ( -24.50) >DroPer_CAF1 10962 97 + 1 CCAUUGGUGUGCUUGGAGC---ACUUUAAAGUCGGCGAUAAUAUUGUCAAGUGUCGACAGUUUUAUCAGCUGCCAGAGCCAAAUGCAAUUUAAGUGCACU .....((((..((((((..---........(.(((((((((.((((((.......)))))).))))).)))).)...((.....))..))))))..)))) ( -28.80) >consensus CCAU_GGU___UUUUAACCACGACUUUAAGUGCGGCGAUAAGGUUGUCAAGUGCGGACAGUUUUAUCAGCCCGCACAG_CAAAUGCAAUUUAAAUGCAUU ....................................(((((..(((((.......)))))..)))))..............((((((.......)))))) (-13.18 = -13.52 + 0.33)



| Location | 14,228,293 – 14,228,388 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 78.92 |
| Mean single sequence MFE | -24.40 |
| Consensus MFE | -10.19 |
| Energy contribution | -9.92 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.42 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.537381 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14228293 95 - 20766785 AAUGCAUUUAAAUUGCAUUUG-CUGUGCGGCCUGAUAAAACUGUCCGCACUUGACAACCUUAUCGUCGCACUUAAAGUCGUGGUUAAAA---ACC-AUGG ((((((.......))))))..-..(((((((..(((((...((((.......))))...)))))))))))).......((((((.....---)))-))). ( -27.30) >DroSim_CAF1 9246 95 - 1 AAUGCAUUUAAAUUGCAUUUG-CCGUACGGCCUGAUAAAACUGUCCGCACUUGACAACCUUAUCGCCGCACUUAAACUCGUGGUUAAAA---ACC-AUGG ((((((.......))))))..-..((.((((..(((((...((((.......))))...))))))))).)).......((((((.....---)))-))). ( -23.70) >DroEre_CAF1 8011 95 - 1 AAUGCAUUUAAAUUGCAUUUG-CUGUGCGGGCUGAUAAAACUGUCCGCACUUGACAACCUUAUCGCCGCACUUAAGCUCGUGGUUAAAA---ACC-AUGC ((((((.......)))))).(-((((((((.(.(((((...((((.......))))...))))))))))))...))).((((((.....---)))-))). ( -28.50) >DroYak_CAF1 7705 87 - 1 AAUGCAUUUAAAUUGCAUUUG---------GCUGAUAAAACUGUCCGCACUUGACAACCUUAUCGGCGCACUUAAACCCGUGGUAAAAA---ACC-AUGC ((((((.......))))))((---------((((((((...((((.......))))...)))))))).))........((((((.....---)))-))). ( -23.00) >DroAna_CAF1 13533 90 - 1 AAUGCAUUUAAAUUGCGUUUC-UUUGGCGG-AAGAUAAACCUGUCGGCACUUGACAACCUUAUCGCAACGCCCAAAGU---GG-AAAAA---ACC-AUUC ((((((.......)))))).(-(((((((.-..(((((...(((((.....)))))...)))))....)).))))))(---((-.....---.))-)... ( -21.70) >DroPer_CAF1 10962 97 - 1 AGUGCACUUAAAUUGCAUUUGGCUCUGGCAGCUGAUAAAACUGUCGACACUUGACAAUAUUAUCGCCGACUUUAAAGU---GCUCCAAGCACACCAAUGG .(((((.......)))))((((...((((....(((((.(.((((((...)))))).).)))))))))........((---((.....)))).))))... ( -22.20) >consensus AAUGCAUUUAAAUUGCAUUUG_CUGUGCGGGCUGAUAAAACUGUCCGCACUUGACAACCUUAUCGCCGCACUUAAACUCGUGGUUAAAA___ACC_AUGC ((((((.......))))))..............(((((...((((.......))))...))))).................................... (-10.19 = -9.92 + -0.28)

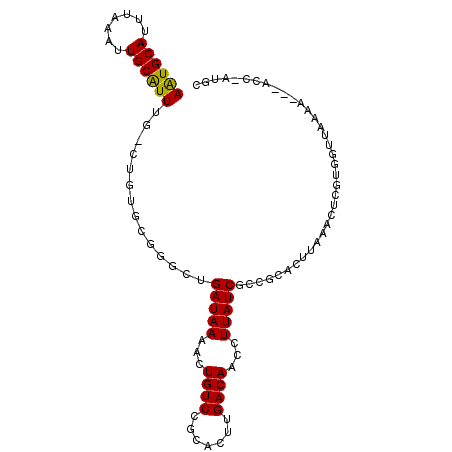
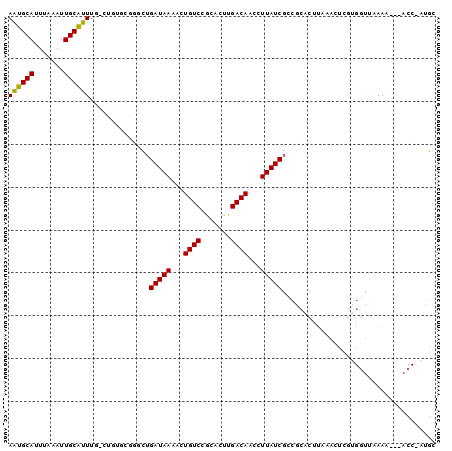
| Location | 14,228,351 – 14,228,460 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 83.87 |
| Mean single sequence MFE | -30.36 |
| Consensus MFE | -18.10 |
| Energy contribution | -18.18 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.60 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.505303 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14228351 109 - 20766785 CCACAUGCGUGUGCUUCGU----UUUAUGGGCACACAUCCUUCGGCUUUCCACGCUCCCC---UGCAUUUUUAUUUU-CGAAUGCAUUUAAAUUGCAUUUGCUGUGCGGCCUGAUAA ........((((((((...----.....)))))))).......((((...(((((.....---.))...........-((((((((.......))))))))..))).))))...... ( -29.30) >DroSec_CAF1 7731 109 - 1 CCACAUGCGUGUGCUUCGU----UUUAUGGGCACACAUCCUUCGGUUUUCCACGCUCCCC---UGCAUUUUUAUUUU-CGAAUGCAUUUAAAUUGCAUUUGCUGUGCGGCCUGAUAA ......((((((((((...----.....)))))))).......((....))..))....(---(((((.........-((((((((.......))))))))..))))))........ ( -27.20) >DroSim_CAF1 9304 109 - 1 CCACAUGCGUGUGCUUCGU----GUUAUGGGCACACAUCCUUCGGUUUUCCACGCUCCCC---UGCAUUUUUAUUUU-CGAAUGCAUUUAAAUUGCAUUUGCCGUACGGCCUGAUAA ...((.((((((((((...----.....))))))))......((((.......((.....---.))...........-.(((((((.......)))))))))))....)).)).... ( -26.80) >DroEre_CAF1 8069 109 - 1 CCACAUGCGUGUGCUUCGU----UUUAUGGGCACACAUCCUUCGGUUUUCCACGCUCCCC---UGCGCUUUUAUUUU-CGAAUGCAUUUAAAUUGCAUUUGCUGUGCGGGCUGAUAA ......((((((((((...----.....)))))))).......((....))..))...((---(((((.........-((((((((.......))))))))..)))))))....... ( -31.60) >DroAna_CAF1 13587 114 - 1 --GGGUGUGGGGGCUCCGGUGUGUUUAUGGCCACACAUCCUUUGGCUUACCGCGCCGCGGAGUGGUGCUUUUAUUUUGUGAAUGCAUUUAAAUUGCGUUUCUUUGGCGG-AAGAUAA --(((((((..((((...(((....))))))).))))))).....(((.((((..((((((((((.....))))))))))((((((.......))))))......))))-))).... ( -36.90) >consensus CCACAUGCGUGUGCUUCGU____UUUAUGGGCACACAUCCUUCGGUUUUCCACGCUCCCC___UGCAUUUUUAUUUU_CGAAUGCAUUUAAAUUGCAUUUGCUGUGCGGCCUGAUAA ....((((((((((.((((.......)))))))))).......((....)).............))))..........((((((((.......))))))))................ (-18.10 = -18.18 + 0.08)

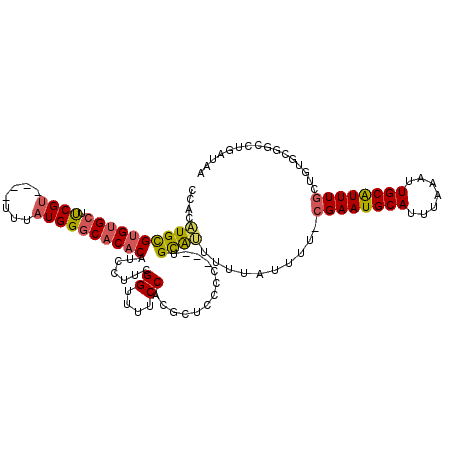
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:17:11 2006