
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,218,274 – 14,218,403 |
| Length | 129 |
| Max. P | 0.854572 |

| Location | 14,218,274 – 14,218,379 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 80.11 |
| Mean single sequence MFE | -49.82 |
| Consensus MFE | -31.76 |
| Energy contribution | -31.90 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.854572 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14218274 105 + 20766785 AGUAC------GGGAUGUGCUGGUGGUUCUAUCACCAGAGGCACUUCAGCAGCCAGCGAAGGAGCAGCAGCGGCCGCUGCCGCUAACCAACCUCUAGACCUGUCCAUGGAU .(.((------(((.......((((((...))))))(((((..((((.((.....)))))).(((.(((((....))))).)))......)))))...))))).)...... ( -37.70) >DroPse_CAF1 44172 111 + 1 ACCACAGGCAGCGGCAGUGGCGGUGGCUCCAUUACACGCGGCACAUCGGCGGCAAGCGAAGGAGCGGCUGCUGCUGCAGCUGCCAAUCAGCCGCUGGAUCUGUCCAUGGAU ......((((((.((((..(((((.(((((.(..(..((.((......)).))..)..).))))).)))))..)))).))))))......(((.((((....))))))).. ( -58.20) >DroEre_CAF1 36879 105 + 1 AGUAC------CGGUGGUGCUGGUGGUUCCAUCACCAGGGGCACUUCAGCCGCCAGCGAAGGGGCAGCGGCGGCCGCCGCUGCCAAUCAGCCUCUGGACCUGUCCAUGGAU (((((------(...))))))((((....)))).((((((((.((((.((.....)))))).(((((((((....))))))))).....)))))))).((.......)).. ( -53.40) >DroYak_CAF1 37704 105 + 1 AGUAC------CGGUGGUGCUGGUGGUUCCAUCACCAGGGGCACUUCGGCCGCCAGCGAAGGAGCGGCAGCGGCUGCCGCUGCCAAUCAACCUCUUGACUUGUCCAUGGAU ....(------((.((((.((((((((...)))))))).(((......))))))).)...(((..((((((((...))))))))..((((....))))....)))..)).. ( -43.30) >DroAna_CAF1 38869 105 + 1 AGCAC------UGGCGGUGGCGGUGGCUCCAUUACCCGGGGCACUUCGGCGGCCAGCGAAGGAGCUGCAGCUGCGGCGGCUGCCAAUCAGCCCCUGGACUUGUCCAUGGAC ..(((------((.(...).)))))..(((((.....(((((.....(((((((..(....).(((((....)))))))))))).....))))).(((....)))))))). ( -48.10) >DroPer_CAF1 43819 111 + 1 ACCACAGGCAGCGGCAGUGGCGGUGGCUCCAUUACACGCGGCACAUCGGCGGCAAGCGAAGGAGCGGCUGCUGCUGCAGCUGCCAAUCAGCCGCUGGAUCUGUCCAUGGAU ......((((((.((((..(((((.(((((.(..(..((.((......)).))..)..).))))).)))))..)))).))))))......(((.((((....))))))).. ( -58.20) >consensus AGCAC______CGGCGGUGCCGGUGGCUCCAUCACCAGGGGCACUUCGGCGGCCAGCGAAGGAGCGGCAGCGGCUGCAGCUGCCAAUCAGCCUCUGGACCUGUCCAUGGAU .((((...........)))).......((((......(((((.((((.((.....))))))..((((((((....))))))))......)))))((((....)))))))). (-31.76 = -31.90 + 0.14)



| Location | 14,218,274 – 14,218,379 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 80.11 |
| Mean single sequence MFE | -45.10 |
| Consensus MFE | -30.66 |
| Energy contribution | -30.08 |
| Covariance contribution | -0.58 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.68 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14218274 105 - 20766785 AUCCAUGGACAGGUCUAGAGGUUGGUUAGCGGCAGCGGCCGCUGCUGCUCCUUCGCUGGCUGCUGAAGUGCCUCUGGUGAUAGAACCACCAGCACAUCCC------GUACU ....((((.(((..((((.((..((..(((((((((....))))))))))).)).))))...)))..((((...(((((.......)))))))))...))------))... ( -39.30) >DroPse_CAF1 44172 111 - 1 AUCCAUGGACAGAUCCAGCGGCUGAUUGGCAGCUGCAGCAGCAGCCGCUCCUUCGCUUGCCGCCGAUGUGCCGCGUGUAAUGGAGCCACCGCCACUGCCGCUGCCUGUGGU ........((((...(((((((....((((.(((((....))))).(((((...((.(((.((......)).))).))...)))))....))))..))))))).))))... ( -47.00) >DroEre_CAF1 36879 105 - 1 AUCCAUGGACAGGUCCAGAGGCUGAUUGGCAGCGGCGGCCGCCGCUGCCCCUUCGCUGGCGGCUGAAGUGCCCCUGGUGAUGGAACCACCAGCACCACCG------GUACU ..((.((((....))))(..((((...(((((((((....)))))))))((.((((..(.(((......))).)..)))).))......))))..)...)------).... ( -50.50) >DroYak_CAF1 37704 105 - 1 AUCCAUGGACAAGUCAAGAGGUUGAUUGGCAGCGGCAGCCGCUGCCGCUCCUUCGCUGGCGGCCGAAGUGCCCCUGGUGAUGGAACCACCAGCACCACCG------GUACU .....(((....((((.((((..((.(((((((((...))))))))).))))))..))))..))).((((((..(((((.(((.....))).)))))..)------))))) ( -46.40) >DroAna_CAF1 38869 105 - 1 GUCCAUGGACAAGUCCAGGGGCUGAUUGGCAGCCGCCGCAGCUGCAGCUCCUUCGCUGGCCGCCGAAGUGCCCCGGGUAAUGGAGCCACCGCCACCGCCA------GUGCU (((....))).((..(.(((((...(((((.(((...((....))(((......)))))).)))))...))))).(((..(((........)))..))).------)..)) ( -40.40) >DroPer_CAF1 43819 111 - 1 AUCCAUGGACAGAUCCAGCGGCUGAUUGGCAGCUGCAGCAGCAGCCGCUCCUUCGCUUGCCGCCGAUGUGCCGCGUGUAAUGGAGCCACCGCCACUGCCGCUGCCUGUGGU ........((((...(((((((....((((.(((((....))))).(((((...((.(((.((......)).))).))...)))))....))))..))))))).))))... ( -47.00) >consensus AUCCAUGGACAGGUCCAGAGGCUGAUUGGCAGCGGCAGCAGCUGCCGCUCCUUCGCUGGCCGCCGAAGUGCCCCGGGUAAUGGAACCACCACCACCGCCG______GUACU .((((((((....))).(((((...(((((.(((((....))))).((......)).....)))))...))))).....)))))........................... (-30.66 = -30.08 + -0.58)
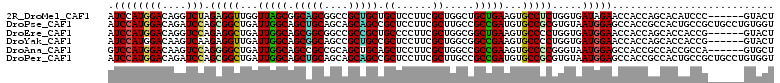
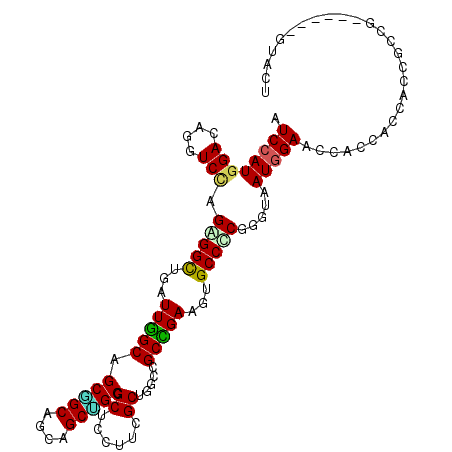
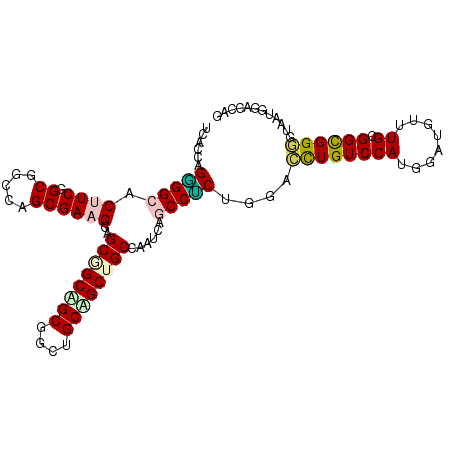
| Location | 14,218,299 – 14,218,403 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 80.90 |
| Mean single sequence MFE | -45.95 |
| Consensus MFE | -31.96 |
| Energy contribution | -31.80 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.543587 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14218299 104 + 20766785 UCACCAGAGGCACUUCAGCAGCCAGCGAAGGAGCAGCAGCGGCCGCUGCCGCUAACCAACCUCUAGACCUGUCCAUGGAUGUUUGCGGCGGGGUAAUGGAUCAG ...(((((((..((((.((.....)))))).(((.(((((....))))).)))......)))))...((((((((........)).)))))).....))..... ( -36.60) >DroPse_CAF1 44203 104 + 1 UUACACGCGGCACAUCGGCGGCAAGCGAAGGAGCGGCUGCUGCUGCAGCUGCCAAUCAGCCGCUGGAUCUGUCCAUGGAUGUCUGCGGCGGAGUCCACGGCGAG .....(((((.(((((((((((...(....).((((((((....))))))))......))))))(((....)))...))))))))))((.(......).))... ( -46.70) >DroEre_CAF1 36904 104 + 1 UCACCAGGGGCACUUCAGCCGCCAGCGAAGGGGCAGCGGCGGCCGCCGCUGCCAAUCAGCCUCUGGACCUGUCCAUGGAUGUUUGCGGCGGGGUAAUAGACCAG ...((((((((.((((.((.....)))))).(((((((((....))))))))).....)))))))).((((((((........)).))))))............ ( -52.50) >DroYak_CAF1 37729 104 + 1 UCACCAGGGGCACUUCGGCCGCCAGCGAAGGAGCGGCAGCGGCUGCCGCUGCCAAUCAACCUCUUGACUUGUCCAUGGAUGUUUGCGGCGGGGGAAUGGACCAG ......((..((((((.(((((.(((.(((....((((((((...))))))))..((((....)))))))(((....)))))).))))).))))..))..)).. ( -40.90) >DroAna_CAF1 38894 104 + 1 UUACCCGGGGCACUUCGGCGGCCAGCGAAGGAGCUGCAGCUGCGGCGGCUGCCAAUCAGCCCCUGGACUUGUCCAUGGACGUGUGUGGUGGCGCCAUGGAGCAG ...((.(((((.....(((((((..(....).(((((....)))))))))))).....))))).))...(((((((((.(((........)))))))))).)). ( -52.30) >DroPer_CAF1 43850 104 + 1 UUACACGCGGCACAUCGGCGGCAAGCGAAGGAGCGGCUGCUGCUGCAGCUGCCAAUCAGCCGCUGGAUCUGUCCAUGGAUGUCUGCGGCGGAGUCCACGGCGAG .....(((((.(((((((((((...(....).((((((((....))))))))......))))))(((....)))...))))))))))((.(......).))... ( -46.70) >consensus UCACCAGGGGCACUUCGGCGGCCAGCGAAGGAGCGGCAGCGGCUGCAGCUGCCAAUCAGCCUCUGGACCUGUCCAUGGAUGUUUGCGGCGGGGUAAUGGACCAG ......(((((.((((.((.....))))))..((((((((....))))))))......)))))....((((((((........)).))))))............ (-31.96 = -31.80 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:17:08 2006