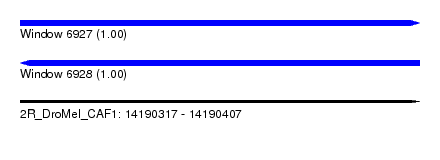
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,190,317 – 14,190,407 |
| Length | 90 |
| Max. P | 0.996974 |

| Location | 14,190,317 – 14,190,407 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 87.31 |
| Mean single sequence MFE | -24.45 |
| Consensus MFE | -21.01 |
| Energy contribution | -22.57 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.22 |
| Mean z-score | -3.91 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.78 |
| SVM RNA-class probability | 0.996974 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
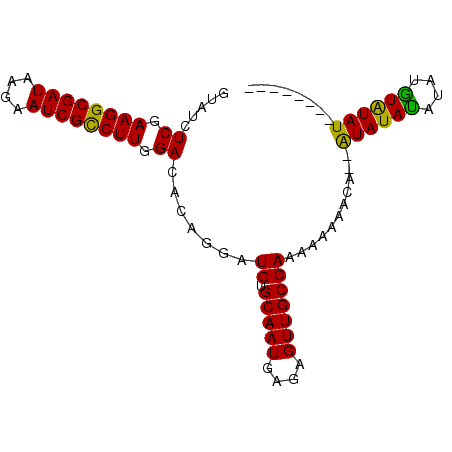
>2R_DroMel_CAF1 14190317 90 + 20766785 GUAUCUCGAAGGCGAUAAGAAUCGUCUUGGACACAGGAUGUGCAAUGAGAGUUGCCAAAAAAAACA--AUAUAUGUACAUAUAUAUAUAAUA .....((.((((((((....)))))))).)).......((.(((((....))))))).........--((((((((....)))))))).... ( -22.70) >DroSec_CAF1 7571 82 + 1 GUAUCUCGAAGGCGAUAAGAAUCGCCUUGGACACAGGAUGUGCAAUGAGAGUUGCCAAAAAAAACA--AUAUAUAUAUGUAUAU-------- .....((.((((((((....)))))))).)).......((.(((((....))))))).........--((((((....))))))-------- ( -22.60) >DroSim_CAF1 7664 82 + 1 GUAUCUCGAAGGCGAUAAGAAUCGCCUUGGACACAGGAUGUGCAAUGAGAGUUGCCAAAAAAAACA--AUAUAUAUAUGUAUAU-------- .....((.((((((((....)))))))).)).......((.(((((....))))))).........--((((((....))))))-------- ( -22.60) >DroEre_CAF1 7951 86 + 1 GUAUCUCGAAGGCGAUAAGAAUCGCCUUGGACACAGGAUGUGCAAUGAGAGUUGCCAAAAUAUAU-GCGUAUACAUAUGUGUAU-----GUA .....((.((((((((....)))))))).)).......((.(((((....))))))).......(-((((((((....))))))-----))) ( -29.90) >consensus GUAUCUCGAAGGCGAUAAGAAUCGCCUUGGACACAGGAUGUGCAAUGAGAGUUGCCAAAAAAAACA__AUAUAUAUAUGUAUAU________ .....((.((((((((....)))))))).)).......((.(((((....)))))))...........((((((....))))))........ (-21.01 = -22.57 + 1.56)


| Location | 14,190,317 – 14,190,407 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 87.31 |
| Mean single sequence MFE | -18.67 |
| Consensus MFE | -16.67 |
| Energy contribution | -16.93 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.85 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.57 |
| SVM RNA-class probability | 0.995424 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
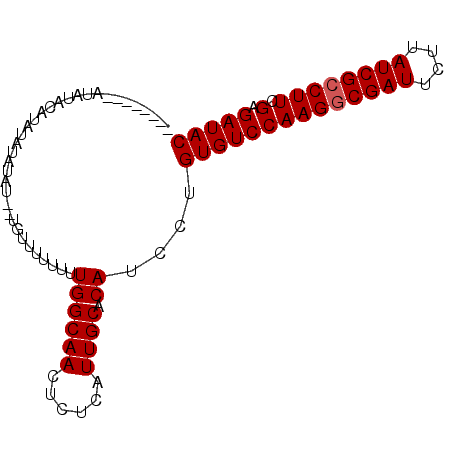
>2R_DroMel_CAF1 14190317 90 - 20766785 UAUUAUAUAUAUAUGUACAUAUAU--UGUUUUUUUUGGCAACUCUCAUUGCACAUCCUGUGUCCAAGACGAUUCUUAUCGCCUUCGAGAUAC .........(((((....)))))(--((((......))))).((((...((((.....))))..(((.((((....)))).))).))))... ( -13.40) >DroSec_CAF1 7571 82 - 1 --------AUAUACAUAUAUAUAU--UGUUUUUUUUGGCAACUCUCAUUGCACAUCCUGUGUCCAAGGCGAUUCUUAUCGCCUUCGAGAUAC --------...............(--((((......))))).((((...((((.....))))..((((((((....)))))))).))))... ( -19.70) >DroSim_CAF1 7664 82 - 1 --------AUAUACAUAUAUAUAU--UGUUUUUUUUGGCAACUCUCAUUGCACAUCCUGUGUCCAAGGCGAUUCUUAUCGCCUUCGAGAUAC --------...............(--((((......))))).((((...((((.....))))..((((((((....)))))))).))))... ( -19.70) >DroEre_CAF1 7951 86 - 1 UAC-----AUACACAUAUGUAUACGC-AUAUAUUUUGGCAACUCUCAUUGCACAUCCUGUGUCCAAGGCGAUUCUUAUCGCCUUCGAGAUAC ...-----......((((((....))-))))....((((((......)))).))....((((((((((((((....)))))))).).))))) ( -21.90) >consensus ________AUAUACAUAUAUAUAU__UGUUUUUUUUGGCAACUCUCAUUGCACAUCCUGUGUCCAAGGCGAUUCUUAUCGCCUUCGAGAUAC ...................................((((((......)))).))....((((((((((((((....)))))))).).))))) (-16.67 = -16.93 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:16:58 2006