| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,188,852 – 14,188,955 |
| Length | 103 |
| Max. P | 0.599894 |

| Location | 14,188,852 – 14,188,955 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 76.66 |
| Mean single sequence MFE | -28.70 |
| Consensus MFE | -13.76 |
| Energy contribution | -14.82 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.599894 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
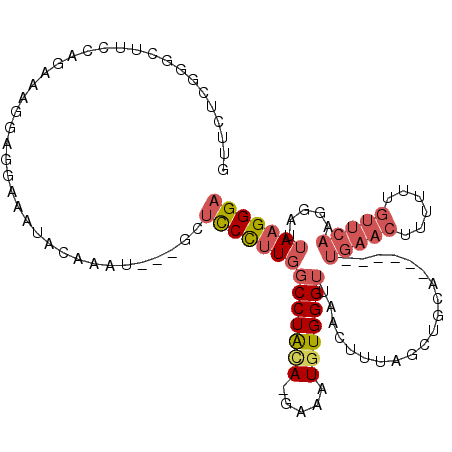
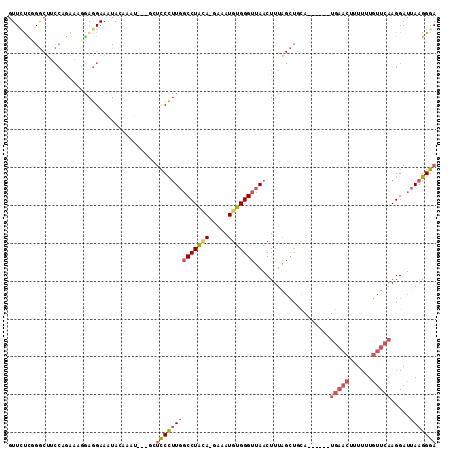
>2R_DroMel_CAF1 14188852 103 + 20766785 GUUCUUUGGCUUCCAGAAUGAAGGAAAUACGAAU---AGUUCCUUGGCCUAUA-GAAAUGUGGGUUAACUUUAGCUGUA------UGAACUUUUUUGUUCAAGGAUUAAGGGA .((((((((..(((.(((..(((((....((.((---((((..((((((((((-....))))))))))....)))))).------))...)))))..)))..))))))))))) ( -26.90) >DroSec_CAF1 6074 103 + 1 GUUAUCUGGCUUCCAGAAAGGAGGAAAUACAAAU---GCUCCCUUGGCCUACA-GAAAUGUGGGUUAACUUUAGCUGCA------UGAACUUUUUUGUUCAAGGAUUAAGGGA ....(((((...)))))..((((.(........)---.)))).((((((((((-....))))))))))((((((((...------(((((......))))).)).)))))).. ( -31.80) >DroSim_CAF1 6113 103 + 1 GUUCUCUGGCUUCCAGAAAGGAGGAAAUACAAAU---GCUCCCUUGGCCUACA-GAAAUGUGGGUUAACUUUAGCUGCA------UGAACUUUUUUGUUCAAGGAUUAAGGGA ....(((((...)))))..((((.(........)---.)))).((((((((((-....))))))))))((((((((...------(((((......))))).)).)))))).. ( -31.60) >DroEre_CAF1 6323 107 + 1 GUUCUUGGGCUGCCAAACAUGAGGAAAUAA-ACU---GAUCCCCUGGCCUACA-GAAAUGUGGGUUAACUUUGGCUGCAUGAGCAUGAACU-UUUUGUUCAAGGAUUAAGGGA ((((....((.((((((.....(((.....-...---..)))..(((((((((-....)))))))))..)))))).))..)))).(((((.-....)))))............ ( -32.80) >DroYak_CAF1 6248 102 + 1 GUUCCCGGGCUGCCAAACAGGAGGAAAUAAGACU---GAUCCCUUGGCCUACA-GAAAUGUGGGUUAACUUUAGCUGCA------UGAACU-UUUUGUUCAAGGAUUAAGGGA ..((((.(((((.....((((.(((.........---..)))))))(((((((-....)))))))......)))))...------(((((.-....)))))........)))) ( -31.60) >DroAna_CAF1 5917 91 + 1 GUUUCCGGA------GGACAAAAGAUCUACAAAGCCAGCAUCUUUUUCCUGAAUCGAAUGUGGGUUAACUUUA----------------CACUUUUAUUCAAGGAUUAAGGGA ...(((..(------(((.(((((((((........)).)))))))))))((((.(((.(((...........----------------)))))).))))..)))........ ( -17.50) >consensus GUUCUCGGGCUUCCAGAAAGGAGGAAAUACAAAU___GCUCCCUUGGCCUACA_GAAAUGUGGGUUAACUUUAGCUGCA______UGAACUUUUUUGUUCAAGGAUUAAGGGA .......................................((((((((((((((.....)))))))....................(((((......))))).....))))))) (-13.76 = -14.82 + 1.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:16:56 2006