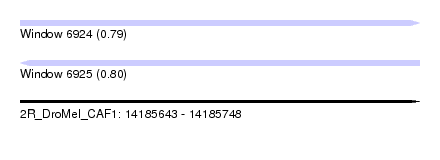
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,185,643 – 14,185,748 |
| Length | 105 |
| Max. P | 0.799413 |

| Location | 14,185,643 – 14,185,748 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.22 |
| Mean single sequence MFE | -20.35 |
| Consensus MFE | -18.58 |
| Energy contribution | -18.75 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.786271 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
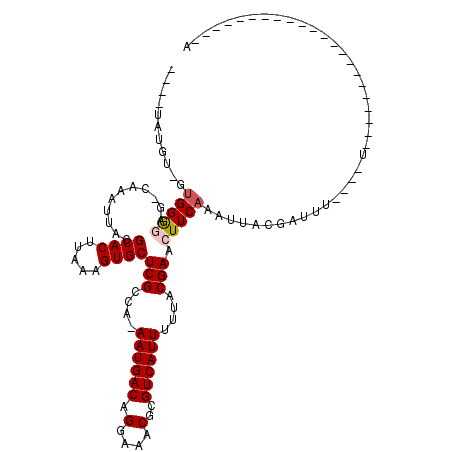
>2R_DroMel_CAF1 14185643 105 + 20766785 UA---A------UACAAAAAAAAAAAAAAAAUCGUAAUUUGAAGUUCGUAAAAAUGACGCGUUUCCUGUCAUU-UGGCGAGCACUUUAAGUGCGUAAUUUG-CUAUCCGCCAAAUA---- ..---.------....................((((.((((((((((((..((((((((.......)))))))-).))))..))))))))))))..(((((-(.....).))))).---- ( -19.70) >DroVir_CAF1 6864 91 + 1 U----------------------A----AAAUCGUAAUUUGAAGUUCGUAAAAAUGACGCGUUUCCUGUCAUU-UGGCGAGCACUUUAAGUGCGUAAUUUG-CUCCCCAC-ACACACUGA .----------------------.----....((((.((((((((((((..((((((((.......)))))))-).))))..)))))))))))).......-........-......... ( -18.80) >DroPse_CAF1 9739 90 + 1 U----------------------A--AAAAAUCGUAAUUUGAAGUUCGUAAAAAUGACGCGUUUCCUGUCAUUUUGGCGAGCACUUUAAGUGCGUAAUUUG-CUAUCCAC-AAAUA---- .----------------------.--......((((.((((((((((((.(((((((((.......))))))))).))))..))))))))))))..(((((-.......)-)))).---- ( -21.90) >DroGri_CAF1 3388 91 + 1 U----------------------A----AAAUCGUAAUUUGAAGUUCGAAAAAAUGACGCGUUUCCUGUCAUU-UGGCGAGCACUUUAAGUGCGUAAUUUG-CUCCCCAC-ACACACACA .----------------------.----....((((.(((((((((((...((((((((.......)))))))-)..)))..)))))))))))).......-........-......... ( -18.70) >DroWil_CAF1 4562 118 + 1 UCGAAAUAUACCUACAGAUACAAAACAAAUCUCGUAAUUUGAAGUUCGUAAAAAUGACGCGUUUCCUGUCAUU-UGGCGAGCACUUUAAGUGCGUAAUUUGGCUCCCCAC-AAAAACACA .........................(((((..((((.((((((((((((..((((((((.......)))))))-).))))..))))))))))))..))))).........-......... ( -24.20) >DroMoj_CAF1 7156 84 + 1 U----------------------A----AAAUCGUAAUUUGAAGUUCGUAAAAAUGACGCGUUUCCUGUCAUU-UGGCGAGCACUUUAAGUGCGUAAUUUG-CUCCCCAC-AC------- .----------------------.----....((((.((((((((((((..((((((((.......)))))))-).))))..)))))))))))).......-........-..------- ( -18.80) >consensus U______________________A____AAAUCGUAAUUUGAAGUUCGUAAAAAUGACGCGUUUCCUGUCAUU_UGGCGAGCACUUUAAGUGCGUAAUUUG_CUCCCCAC_AAACA____ ................................((((.((((((((((((...(((((((.......)))))))...))))..)))))))))))).......................... (-18.58 = -18.75 + 0.17)

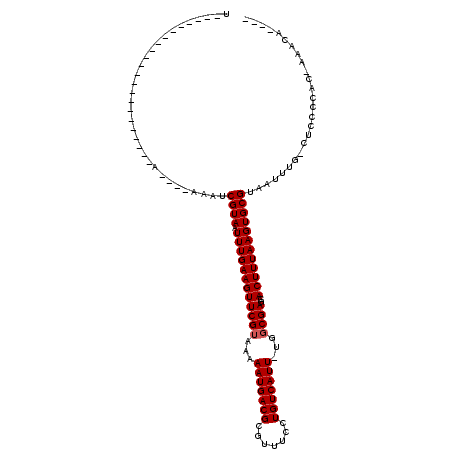
| Location | 14,185,643 – 14,185,748 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.22 |
| Mean single sequence MFE | -23.02 |
| Consensus MFE | -17.75 |
| Energy contribution | -18.03 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.799413 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
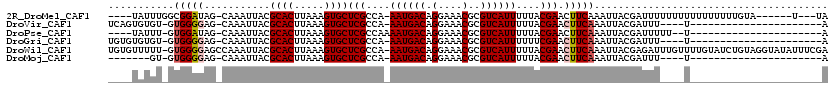
>2R_DroMel_CAF1 14185643 105 - 20766785 ----UAUUUGGCGGAUAG-CAAAUUACGCACUUAAAGUGCUCGCCA-AAUGACAGGAAACGCGUCAUUUUUACGAACUUCAAAUUACGAUUUUUUUUUUUUUUUUGUA------U---UA ----.(((((((((((..-...)))..((((.....)))).)))))-)))(((.(....)..)))...................(((((..............)))))------.---.. ( -20.74) >DroVir_CAF1 6864 91 - 1 UCAGUGUGU-GUGGGGAG-CAAAUUACGCACUUAAAGUGCUCGCCA-AAUGACAGGAAACGCGUCAUUUUUACGAACUUCAAAUUACGAUUU----U----------------------A ..(((.(((-(.((.(((-((..(((......)))..))))).))(-((((((.(....)..))))))).)))).)))..............----.----------------------. ( -23.70) >DroPse_CAF1 9739 90 - 1 ----UAUUU-GUGGAUAG-CAAAUUACGCACUUAAAGUGCUCGCCAAAAUGACAGGAAACGCGUCAUUUUUACGAACUUCAAAUUACGAUUUUU--U----------------------A ----.((((-(.((....-........((((.....))))(((..((((((((.(....)..))))))))..))).)).)))))..........--.----------------------. ( -21.10) >DroGri_CAF1 3388 91 - 1 UGUGUGUGU-GUGGGGAG-CAAAUUACGCACUUAAAGUGCUCGCCA-AAUGACAGGAAACGCGUCAUUUUUUCGAACUUCAAAUUACGAUUU----U----------------------A .(((..((.-((((.(((-((..(((......)))..))))).))(-((((((.(....)..)))))))......))..))...))).....----.----------------------. ( -22.90) >DroWil_CAF1 4562 118 - 1 UGUGUUUUU-GUGGGGAGCCAAAUUACGCACUUAAAGUGCUCGCCA-AAUGACAGGAAACGCGUCAUUUUUACGAACUUCAAAUUACGAGAUUUGUUUUGUAUCUGUAGGUAUAUUUCGA (((((....-.(((....)))....)))))......(((((..(.(-((((((.(....)..))))))).((((((...((((((....)))))).))))))...)..)))))....... ( -28.20) >DroMoj_CAF1 7156 84 - 1 -------GU-GUGGGGAG-CAAAUUACGCACUUAAAGUGCUCGCCA-AAUGACAGGAAACGCGUCAUUUUUACGAACUUCAAAUUACGAUUU----U----------------------A -------((-(.((.(((-((..(((......)))..))))).))(-((((((.(....)..))))))).)))...................----.----------------------. ( -21.50) >consensus ____UAUGU_GUGGGGAG_CAAAUUACGCACUUAAAGUGCUCGCCA_AAUGACAGGAAACGCGUCAUUUUUACGAACUUCAAAUUACGAUUU____U______________________A ...........(((((...........((((.....))))(((....((((((.(....)..))))))....))).)))))....................................... (-17.75 = -18.03 + 0.28)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:16:55 2006