| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,178,554 – 14,178,658 |
| Length | 104 |
| Max. P | 0.821947 |

| Location | 14,178,554 – 14,178,658 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.96 |
| Mean single sequence MFE | -23.98 |
| Consensus MFE | -14.76 |
| Energy contribution | -15.82 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.821947 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
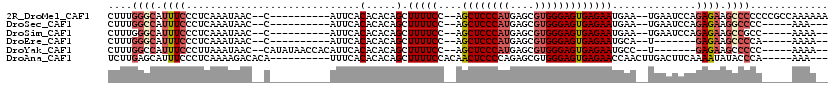
>2R_DroMel_CAF1 14178554 104 + 20766785 CUUUGGGCAUUUCCCUCAAAUAAC--C----------AUUCACACACAGCUUUUCC--AGCUCCCAUGAGCGUGGGAGUGAGAAUGAA--UGAAUCCAGAGAAGCCCCCCCGCCAAAAAA ....((((.((((...........--(----------((((.(((...(((.....--)))((((((....))))))))).)))))..--((....)))))).))))............. ( -28.00) >DroSec_CAF1 22760 96 + 1 CUUUGGCCAUUUCCCUCAAAUAAC--C----------AUUCACACACAGCUUUUCC--AGCUCCCAUGAGCGUGGGAGUGAGAAUGAA--UGAAUCCAGAGAAGGCCCC-----AAA--- ....((((.((((...........--(----------((((.(((...(((.....--)))((((((....))))))))).)))))..--((....)))))).))))..-----...--- ( -28.00) >DroSim_CAF1 22921 97 + 1 CUUUGGGCAUUUCCCUCAAAUAAC--C----------AUUCACACACAGCUUUUCC--AGCUCCCAUGAGCGUGGGAGUGAGAAUGAA--UGAAUCCAGAGAAGCCGCC-----AAAA-- .....(((.((((...........--(----------((((.(((...(((.....--)))((((((....))))))))).)))))..--((....)))))).)))...-----....-- ( -25.60) >DroEre_CAF1 23676 90 + 1 CUUUGGGCAUUUCCCUCAAAUAAC--C----------AUUCACACACAGCUUUUCC--AGCUCCCAUGAGCGUGGGAGUGAGAAUGCA--U-------GAGAAGCCCCA-----AAAA-- .((((((.......((((......--(----------((((.(((...(((.....--)))((((((....))))))))).)))))..--)-------)))....))))-----))..-- ( -26.00) >DroYak_CAF1 23221 100 + 1 CUUUGGCCAUUUCCCUUAAAUAAC--CAUAUAACCACAUUCACACACAGCUUUUCC--AGCUCCCAUGAGCGUGGGAGUGAGAAUGCC--U-------GAGAAGCCCCC-----AAAA-- .(((((...((((......(((..--..)))...............(((...(((.--.((((((((....))))))))..)))...)--)-------).))))...))-----))).-- ( -21.70) >DroAna_CAF1 24487 102 + 1 UCUUGAGCAUUUCCCUCAAAAGACACA----------UUUCACACACAGCUUUUCCACAACUCCCCAGAGCGUGGGAGUGAGAACCAACUUGACUUCAAAAUAUACCCA-----AAA--- ..(((((.......)))))........----------((((((.........((((((..(((....))).))))))))))))..........................-----...--- ( -14.60) >consensus CUUUGGGCAUUUCCCUCAAAUAAC__C__________AUUCACACACAGCUUUUCC__AGCUCCCAUGAGCGUGGGAGUGAGAAUGAA__UGAAUCCAGAGAAGCCCCC_____AAAA__ ....((((.((((.............................(.....).(((((....((((((((....)))))))))))))..............)))).))))............. (-14.76 = -15.82 + 1.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:16:50 2006