| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 14,177,126 – 14,177,247 |
| Length | 121 |
| Max. P | 0.551511 |

| Location | 14,177,126 – 14,177,227 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 75.32 |
| Mean single sequence MFE | -24.82 |
| Consensus MFE | -13.25 |
| Energy contribution | -12.87 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.551511 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
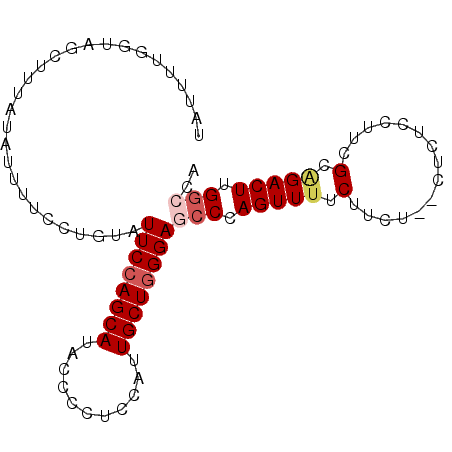
>2R_DroMel_CAF1 14177126 101 - 20766785 UUC-CUGUAUUCCAGCAUACCCCUCCAUUGCUGGGAGCC-----CAGUUUUCUUCU--CUCUCCUUCGCAGACUUGGCCACACACAACUCGACGUCUUCAGCUGGCGA----U ...-.(((.((((((((...........))))))))(((-----.(((((.(....--.........).))))).))).....)))......((((.......)))).----. ( -21.92) >DroSec_CAF1 21273 100 - 1 UUC-CUGUAUUCCAGCAUACCCCUCCAUUGCUGGGAGCC-----CAGUUUUCUUCU--CUCUCCUUCGCGGACUUGGCCACACACAA-UCGCCGUCUUCAGCUGGCGA----U ...-.(((.((((((((...........))))))))(((-----.((((..(....--.........)..)))).))).....)))(-((((((.(....).))))))----) ( -27.02) >DroSim_CAF1 21452 101 - 1 UUC-CUGUAUUCCAGCAUACCCCUUCAUUGCUGGGAGCC-----CAGUUUUCUUCU--CUCUCCUUCGCAGACUUGGCCACACACAACUCGCCGUCUUCAGCUGGCGA----U ...-.(((.((((((((...........))))))))(((-----.(((((.(....--.........).))))).))).....)))..((((((.(....).))))))----. ( -25.92) >DroEre_CAF1 22177 99 - 1 AUC-CUGGAUUCCAGCAUACCCUUGCAUUGCUAGGAACC-----CAGUUUUCCUCCCCC----CUCCGCAGACUUGGCCACACACAACUCGCCGUCUUCAGCUGGCGA----U ...-((((.((((((((...........)))).)))).)-----)))............----.........................((((((.(....).))))))----. ( -22.40) >DroYak_CAF1 21827 103 - 1 AUC-CCGAAUUCCAGCAUACCCCUGCAUUGCUGGGAAGC-----CAGUUCUCCUCCCCCUCUCCCCCGCAGACUUGGCCACACACAACUCGCCGUCUUCAGCUGGCGA----U ...-.....((((((((...........)))))))).((-----(((.(((.(..............).))).)))))..........((((((.(....).))))))----. ( -25.64) >DroAna_CAF1 22471 102 - 1 AACUCCAAAGUCCAGCAUACCCUGGGAGCUCUAGGACCUGGUGGAGGUUUUUCU--------GCUUA-AGUUUUUGGCCACACACCGACCGCUG--CUCCAUUGGCAACAAAG ............(((......)))(((((....((...((((((.((((.....--------((...-.))....)))).).))))).))...)--)))).(((....))).. ( -26.00) >consensus AUC_CUGUAUUCCAGCAUACCCCUGCAUUGCUGGGAGCC_____CAGUUUUCCUCU__CUCUCCUUCGCAGACUUGGCCACACACAACUCGCCGUCUUCAGCUGGCGA____U ...........(((((....(((.........)))................................(.((((..(((............))))))).).)))))........ (-13.25 = -12.87 + -0.39)


| Location | 14,177,155 – 14,177,247 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 84.80 |
| Mean single sequence MFE | -17.75 |
| Consensus MFE | -12.14 |
| Energy contribution | -13.02 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.68 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 14177155 92 - 20766785 UCUUUUGGUAGCUUUAUAUUUUCCUGUAUUCCAGCAUACCCCUCCAUUGCUGGGAGCCCAGUUUUCUUCU--CUCUCCUUCGCAGACUUGGCCA ......(((((((..((((......))))...))).))))...(((((((.(((((...((.......))--..)))))..))))...)))... ( -17.50) >DroSec_CAF1 21301 92 - 1 UAUUUUGGUAGCUUUAUAUUUUCCUGUAUUCCAGCAUACCCCUCCAUUGCUGGGAGCCCAGUUUUCUUCU--CUCUCCUUCGCGGACUUGGCCA .....((((..............(((..((((((((...........))))))))...))).........--...(((.....)))....)))) ( -18.70) >DroSim_CAF1 21481 92 - 1 UAUUUUGGUAGCUUUAUAUUUUCCUGUAUUCCAGCAUACCCCUUCAUUGCUGGGAGCCCAGUUUUCUUCU--CUCUCCUUCGCAGACUUGGCCA ......(((((((..((((......))))...))).))))((....((((.(((((...((.......))--..)))))..))))....))... ( -17.50) >DroEre_CAF1 22206 90 - 1 UGUCUUAGUUGCCUCAUAUUAUCCUGGAUUCCAGCAUACCCUUGCAUUGCUAGGAACCCAGUUUUCCUCCCCC----CUCCGCAGACUUGGCCA .(((..((((((........(..((((.((((((((...........)))).)))).))))..).........----....))).))).))).. ( -17.50) >DroYak_CAF1 21856 94 - 1 UAUUACAGUUGCCUUAUAUUAUCCCGAAUUCCAGCAUACCCCUGCAUUGCUGGGAAGCCAGUUCUCCUCCCCCUCUCCCCCGCAGACUUGGCCA ............................((((((((...........)))))))).(((((.(((.(..............).))).))))).. ( -17.54) >consensus UAUUUUGGUAGCUUUAUAUUUUCCUGUAUUCCAGCAUACCCCUCCAUUGCUGGGAGCCCAGUUUUCUUCU__CUCUCCUUCGCAGACUUGGCCA ............................((((((((...........))))))))(((.(((((.(...............).))))).))).. (-12.14 = -13.02 + 0.88)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:16:48 2006